Related Research Articles

An infection is the invasion of tissues by pathogens, their multiplication, and the reaction of host tissues to the infectious agent and the toxins they produce. An infectious disease, also known as a transmissible disease or communicable disease, is an illness resulting from an infection.

An epidemic is the rapid spread of disease to a large number of hosts in a given population within a short period of time. For example, in meningococcal infections, an attack rate in excess of 15 cases per 100,000 people for two consecutive weeks is considered an epidemic.

Herd immunity is a form of indirect protection that applies only to contagious diseases. It occurs when a sufficient percentage of a population has become immune to an infection, whether through previous infections or vaccination, thereby reducing the likelihood of infection for individuals who lack immunity.
In medicine, public health, and biology, transmission is the passing of a pathogen causing communicable disease from an infected host individual or group to a particular individual or group, regardless of whether the other individual was previously infected. The term strictly refers to the transmission of microorganisms directly from one individual to another by one or more of the following means:
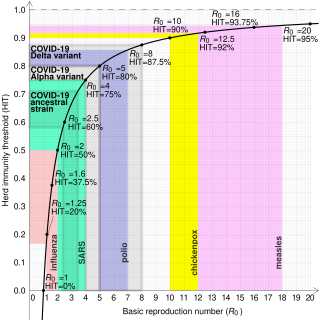
In epidemiology, the basic reproduction number, or basic reproductive number, denoted , of an infection is the expected number of cases directly generated by one case in a population where all individuals are susceptible to infection. The definition assumes that no other individuals are infected or immunized. Some definitions, such as that of the Australian Department of Health, add the absence of "any deliberate intervention in disease transmission". The basic reproduction number is not necessarily the same as the effective reproduction number , which is the number of cases generated in the current state of a population, which does not have to be the uninfected state. is a dimensionless number and not a time rate, which would have units of time−1, or units of time like doubling time.

In epidemiology, an infection is said to be endemic in a specific population or populated place when that infection is constantly present, or maintained at a baseline level, without extra infections being brought into the group as a result of travel or similar means. The term describes the distribution (spread) of an infectious disease among a group of people or within a populated area. An endemic disease always has a steady, predictable number of people getting sick, but that number can be high (hyperendemic) or low (hypoendemic), and the disease can be severe or mild. Also, a disease that is usually endemic can become epidemic.
Mathematical models can project how infectious diseases progress to show the likely outcome of an epidemic and help inform public health and plant health interventions. Models use basic assumptions or collected statistics along with mathematics to find parameters for various infectious diseases and use those parameters to calculate the effects of different interventions, like mass vaccination programs. The modelling can help decide which intervention(s) to avoid and which to trial, or can predict future growth patterns, etc.
Compartmental models are a very general modelling technique. They are often applied to the mathematical modelling of infectious diseases. The population is assigned to compartments with labels – for example, S, I, or R,. People may progress between compartments. The order of the labels usually shows the flow patterns between the compartments; for example SEIS means susceptible, exposed, infectious, then susceptible again.

In infectious disease ecology and epidemiology, a natural reservoir, also known as a disease reservoir or a reservoir of infection, is the population of organisms or the specific environment in which an infectious pathogen naturally lives and reproduces, or upon which the pathogen primarily depends for its survival. A reservoir is usually a living host of a certain species, such as an animal or a plant, inside of which a pathogen survives, often without causing disease for the reservoir itself. By some definitions a reservoir may also be an environment external to an organism, such as a volume of contaminated air or water.
An emergent virus is a virus that is either newly appeared, notably increasing in incidence/geographic range or has the potential to increase in the near future. Emergent viruses are a leading cause of emerging infectious diseases and raise public health challenges globally, given their potential to cause outbreaks of disease which can lead to epidemics and pandemics. As well as causing disease, emergent viruses can also have severe economic implications. Recent examples include the SARS-related coronaviruses, which have caused the 2002-2004 outbreak of SARS (SARS-CoV-1) and the 2019–21 pandemic of COVID-19 (SARS-CoV-2). Other examples include the human immunodeficiency virus which causes HIV/AIDS; the viruses responsible for Ebola; the H5N1 influenza virus responsible for avian flu; and H1N1/09, which caused the 2009 swine flu pandemic. Viral emergence in humans is often a consequence of zoonosis, which involves a cross-species jump of a viral disease into humans from other animals. As zoonotic viruses exist in animal reservoirs, they are much more difficult to eradicate and can therefore establish persistent infections in human populations.

A subclinical infection—sometimes called a preinfection or inapparent infection—is an infection by a pathogen that causes few or no signs or symptoms of infection in the host. Subclinical infections can occur in both humans and animals. Depending on the pathogen, which can be a virus or intestinal parasite, the host may be infectious and able to transmit the pathogen without ever developing symptoms; such a host is called an asymptomatic carrier. Many pathogens, including HIV, typhoid fever, and coronaviruses such as COVID-19 spread in their host populations through subclinical infection.
The eradication of infectious diseases is the reduction of the prevalence of an infectious disease in the global host population to zero.

Airborne transmission or aerosol transmission is transmission of an infectious disease through small particles suspended in the air. Infectious diseases capable of airborne transmission include many of considerable importance both in human and veterinary medicine. The relevant infectious agent may be viruses, bacteria, or fungi, and they may be spread through breathing, talking, coughing, sneezing, raising of dust, spraying of liquids, flushing toilets, or any activities which generate aerosol particles or droplets. This is the transmission of diseases via transmission of an infectious agent, and does not include diseases caused by air pollution.
Viral phylodynamics is defined as the study of how epidemiological, immunological, and evolutionary processes act and potentially interact to shape viral phylogenies. Since the coining of the term in 2004, research on viral phylodynamics has focused on transmission dynamics in an effort to shed light on how these dynamics impact viral genetic variation. Transmission dynamics can be considered at the level of cells within an infected host, individual hosts within a population, or entire populations of hosts.

A superspreading event (SSEV) is an event in which an infectious disease is spread much more than usual, while an unusually contagious organism infected with a disease is known as a superspreader. In the context of a human-borne illness, a superspreader is an individual who is more likely to infect others, compared with a typical infected person. Such superspreaders are of particular concern in epidemiology.
Cross-species transmission (CST), also called interspecies transmission, host jump, or spillover, is the transmission of an infectious pathogen, such as a virus, between hosts belonging to different species. Once introduced into an individual of a new host species, the pathogen may cause disease for the new host and/or acquire the ability to infect other individuals of the same species, allowing it to spread through the new host population. The phenomenon is most commonly studied in virology, but cross-species transmission may also occur with bacterial pathogens or other types of microorganisms.

Ira M. Longini is an American biostatistician and infectious disease epidemiologist.
Groups of animals and humans that live in places with high population density have an increased risk of disease prevalence. In looking at sociality and disease transmission, an examination of how social grouping strategies may reduce or increase the spread of disease is critical for the health of large groups of people. Social groups, community structures, and cultures affect the use of different strategies and behaviors to reduce the spread of disease.
Disease ecology is a sub-discipline of ecology concerned with the mechanisms, patterns, and effects of host-pathogen interactions, particularly those of infectious diseases. For example, it examines how parasites spread through and influence wildlife populations and communities. By studying the flow of diseases within the natural environment, scientists seek to better understand how changes within our environment can shape how pathogens, and other diseases, travel. Therefore, diseases ecology seeks to understand the links between ecological interactions and disease evolution. New emerging and re-emerging infectious diseases are increasing at unprecedented rates which can have lasting impacts on public health, ecosystem health, and biodiversity.
In the field of epidemiology, source attribution refers to a category of methods with the objective of reconstructing the transmission of an infectious disease from a specific source, such as a population, individual, or location. For example, source attribution methods may be used to trace the origin of a new pathogen that recently crossed from another host species into humans, or from one geographic region to another. It may be used to determine the common source of an outbreak of a foodborne infectious disease, such as a contaminated water supply. Finally, source attribution may be used to estimate the probability that an infection was transmitted from one specific individual to another, i.e., "who infected whom".
References
- 1 2 Bartlett, M. S. (1960). "The critical community size for measles in the United States". Journal of the Royal Statistical Society. Series A (General). 123 (1): 37–44. doi:10.2307/2343186. JSTOR 2343186.
- ↑ Haydon, Daniel T; Cleaveland, Sarah; Taylor, Louise H; Laurenson, M Karen (2002). "Identifying reservoirs of infection: a conceptual and practical challenge". Emerging Infectious Diseases. 8 (12): 1468–1473. doi:10.3201/eid0812.010317. PMC 2738515 . PMID 12498665.