Related Research Articles

In genetics, the phenotype is the set of observable characteristics or traits of an organism. The term covers the organism's morphology, its developmental processes, its biochemical and physiological properties, its behavior, and the products of behavior. An organism's phenotype results from two basic factors: the expression of an organism's genetic code and the influence of environmental factors. Both factors may interact, further affecting the phenotype. When two or more clearly different phenotypes exist in the same population of a species, the species is called polymorphic. A well-documented example of polymorphism is Labrador Retriever coloring; while the coat color depends on many genes, it is clearly seen in the environment as yellow, black, and brown. Richard Dawkins in 1978 and then again in his 1982 book The Extended Phenotype suggested that one can regard bird nests and other built structures such as caddisfly larva cases and beaver dams as "extended phenotypes".
Nature versus nurture is a long-standing debate in biology and society about the relative influence on human beings of their genetic inheritance (nature) and the environmental conditions of their development (nurture). The alliterative expression "nature and nurture" in English has been in use since at least the Elizabethan period and goes back to medieval French. The complementary combination of the two concepts is an ancient concept. Nature is what people think of as pre-wiring and is influenced by genetic inheritance and other biological factors. Nurture is generally taken as the influence of external factors after conception e.g. the product of exposure, experience and learning on an individual.

A generegulatory network (GRN) is a collection of molecular regulators that interact with each other and with other substances in the cell to govern the gene expression levels of mRNA and proteins which, in turn, determine the function of the cell. GRN also play a central role in morphogenesis, the creation of body structures, which in turn is central to evolutionary developmental biology (evo-devo).

A protein isoform, or "protein variant", is a member of a set of highly similar proteins that originate from a single gene or gene family and are the result of genetic differences. While many perform the same or similar biological roles, some isoforms have unique functions. A set of protein isoforms may be formed from alternative splicings, variable promoter usage, or other post-transcriptional modifications of a single gene; post-translational modifications are generally not considered. Through RNA splicing mechanisms, mRNA has the ability to select different protein-coding segments (exons) of a gene, or even different parts of exons from RNA to form different mRNA sequences. Each unique sequence produces a specific form of a protein.

Drosophila embryogenesis, the process by which Drosophila embryos form, is a favorite model system for genetics and developmental biology. The study of its embryogenesis unlocked the century-long puzzle of how development was controlled, creating the field of evolutionary developmental biology. The small size, short generation time, and large brood size make it ideal for genetic studies. Transparent embryos facilitate developmental studies. Drosophila melanogaster was introduced into the field of genetic experiments by Thomas Hunt Morgan in 1909.
Evolvability is defined as the capacity of a system for adaptive evolution. Evolvability is the ability of a population of organisms to not merely generate genetic diversity, but to generate adaptive genetic diversity, and thereby evolve through natural selection.

Caulobacter crescentus is a Gram-negative, oligotrophic bacterium widely distributed in fresh water lakes and streams. The taxon is more properly known as Caulobacter vibrioides.
In genetics, expressivity is the degree to which a phenotype is expressed by individuals having a particular genotype. Alternatively, it may refer to the expression of a particular gene by individuals having a certain phenotype. Expressivity is related to the intensity of a given phenotype; it differs from penetrance, which refers to the proportion of individuals with a particular genotype that share the same phenotype.

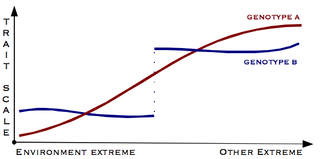
Gene–environment interaction is when two different genotypes respond to environmental variation in different ways. A norm of reaction is a graph that shows the relationship between genes and environmental factors when phenotypic differences are continuous. They can help illustrate GxE interactions. When the norm of reaction is not parallel, as shown in the figure below, there is a gene by environment interaction. This indicates that each genotype responds to environmental variation in a different way. Environmental variation can be physical, chemical, biological, behavior patterns or life events.

Copy number variation (CNV) is a phenomenon in which sections of the genome are repeated and the number of repeats in the genome varies between individuals. Copy number variation is a type of structural variation: specifically, it is a type of duplication or deletion event that affects a considerable number of base pairs. Approximately two-thirds of the entire human genome may be composed of repeats and 4.8–9.5% of the human genome can be classified as copy number variations. In mammals, copy number variations play an important role in generating necessary variation in the population as well as disease phenotype.
Canalisation is a measure of the ability of a population to produce the same phenotype regardless of variability of its environment or genotype. It is a form of evolutionary robustness. The term was coined in 1942 by C. H. Waddington to capture the fact that "developmental reactions, as they occur in organisms submitted to natural selection...are adjusted so as to bring about one definite end-result regardless of minor variations in conditions during the course of the reaction". He used this word rather than robustness to consider that biological systems are not robust in quite the same way as, for example, engineered systems.

A nuclear gene is a gene that has its DNA nucleotide sequence physically situated within the cell nucleus of a eukaryotic organism. This term is employed to differentiate nuclear genes, which are located in the cell nucleus, from genes that are found in mitochondria or chloroplasts. The vast majority of genes in eukaryotes are nuclear.
Transcriptional bursting, also known as transcriptional pulsing, is a fundamental property of genes in which transcription from DNA to RNA can occur in "bursts" or "pulses", which has been observed in diverse organisms, from bacteria to mammals.
Transcriptional noise is a primary cause of the variability (noise) in gene expression occurring between cells in isogenic populations. A proposed source of transcriptional noise is transcriptional bursting although other sources of heterogeneity, such as unequal separation of cell contents at mitosis are also likely to contribute considerably. Bursting transcription, as opposed to simple probabilistic models of transcription, reflects multiple states of gene activity, with fluctuations between states separated by irregular intervals, generating uneven protein expression between cells. Noise in gene expression can have tremendous consequences on cell behaviour, and must be mitigated or integrated. In certain contexts, such as establishment of viral latency, the survival of microbes in rapidly changing stressful environments, or several types of scattered differentiation, the variability may be essential. Variability also impacts upon the effectiveness of clinical treatment, with resistance of bacteria and yeast to antibiotics demonstrably caused by non-genetic differences. Variability in gene expression may also contribute to resistance of sub-populations of cancer cells to chemotherapy and appears to be a barrier to curing HIV.

In evolutionary biology, robustness of a biological system is the persistence of a certain characteristic or trait in a system under perturbations or conditions of uncertainty. Robustness in development is known as canalization. According to the kind of perturbation involved, robustness can be classified as mutational, environmental, recombinational, or behavioral robustness etc. Robustness is achieved through the combination of many genetic and molecular mechanisms and can evolve by either direct or indirect selection. Several model systems have been developed to experimentally study robustness and its evolutionary consequences.
Cellular noise is random variability in quantities arising in cellular biology. For example, cells which are genetically identical, even within the same tissue, are often observed to have different expression levels of proteins, different sizes and structures. These apparently random differences can have important biological and medical consequences.
Johan Paulsson is a Swedish mathematician and systems biologist at Harvard Medical School. He is a leading researcher in systems biology and stochastic processes, specializing in stochasticity in gene networks and plasmid reproduction.

In cell biology, single-cell variability occurs when individual cells in an otherwise similar population differ in shape, size, position in the cell cycle, or molecular-level characteristics. Such differences can be detected using modern single-cell analysis techniques. Investigation of variability within a population of cells contributes to understanding of developmental and pathological processes,
State switching is a fundamental physiological process in which a cell/organism undergoes spontaneous, and potentially reversible, transitions between different phenotypes. Thus, the ability to switch states/phenotypes is a key feature of development and normal function of cells within most multicellular organisms that enables the cell to respond to various intrinsic and extrinsic cues and stimuli in a concerted fashion enabling them to ‘make’ appropriate cellular decisions. Although state switching is essential for normal functioning, the repertoire of phenotypes in a normal cell is albeit limited.
Harley H. McAdams is an American physicist, microbial geneticist, and developmental biologist. McAdams and his collaborators have published foundational insights on the nature of genetic regulatory logic and cell biology, the molecular basis for inevitable random variation levels of protein production between different cells, and genetic logic circuits that control the bacterial cell cycle. McAdams is married to Lucy Shapiro. They were jointly awarded the 2009 John Scott Medal for “bringing the methods of electrical circuit analysis to the description of genetic networks of the simple bacterium Caulobacter.”
References
- ↑ Yampolsky LY, Scheiner SR (1994). "Developmental Noise, Phenotypic Plasticity, and Allozyme Heterozygosity in Daphnia". Evolution. 48 (5): 1715–22. doi:10.2307/2410259. JSTOR 2410259. PMID 28568411.
- ↑ Kærn, Mads; Elston, Timothy C.; Blake, William J.; Collins, James J. (June 2005). "Stochasticity in gene expression: from theories to phenotypes". Nature Reviews Genetics. 6 (6): 451–464. doi:10.1038/nrg1615. ISSN 1471-0064. PMID 15883588. S2CID 1028111.
- ↑ Horikawa K, Ishimatsu K, Yoshimoto E, Kondo S, Takeda H (June 2006). "Noise-resistant and synchronized oscillation of the segmentation clock". Nature. 441 (7094): 719–23. Bibcode:2006Natur.441..719H. doi:10.1038/nature04861. PMID 16760970. S2CID 4372075.
- ↑ Raser, J. M.; O'Shea, E. K. (2005). "Noise in gene expression: origins, consequences, and control". Science . 309 (5743): 2010–2013. Bibcode:2005Sci...309.2010R. doi:10.1126/science.1105891. PMC 1360161 . PMID 16179466.
- ↑ "Do identical twins have identical fingerprints?". Washington State Twin Registry. 1 October 2015. Retrieved 29 September 2019.
- ↑ Barkai, N.; Shilo, B. Z. (2007). "Variability and robustness in biomolecular systems". Mol Cell. 28 (5): 755–760. doi: 10.1016/j.molcel.2007.11.013 . PMID 18082601.
- ↑ Kussell, E.; Kishony, R.; Balaban, N. Q.; Leibler, S. (2005). "Bacterial persistence: a model of survival in changing environments". Genetics. 169 (4): 1807–1814. doi:10.1534/genetics.104.035352. PMC 1449587 . PMID 15687275.
- ↑ Chapman-McQuiston, E. (2007). The Effect of Noisy Protein Expression on E. coli/Phage Dynamics (Thesis).
- ↑ Blake, W. J.; et al. (2006). "Phenotypic consequences of promoter-mediated transcriptional noise". Mol Cell. 24 (6): 853–865. doi: 10.1016/j.molcel.2006.11.003 . PMID 17189188.
- 1 2 Wernet, M. F.; et al. (2006). "Stochastic spineless expression creates the retinal mosaic for colour vision". Nature . 440 (7081): 174–180. Bibcode:2006Natur.440..174W. doi:10.1038/nature04615. PMC 3826883 . PMID 16525464.
- 1 2 Forde, B. G. (2009). "Is it good noise? The role of developmental instability in the shaping of a root system". J Exp Bot. 60 (14): 3989–4002. doi: 10.1093/jxb/erp265 . PMID 19759097.
- ↑ Balázsi, Gábor; van Oudenaarden, Alexander; Collins, James J (2011). "Cellular Decision Making and Biological Noise: From Microbes to Mammals". Cell. 144 (6): 910–925. doi:10.1016/j.cell.2011.01.030. PMC 3068611 . PMID 21414483.
- 1 2 Ishimatsu, K.; Horikawa, K.; Takeda, H. (2007). "Coupling cellular oscillators: a mechanism that maintains synchrony against developmental noise in the segmentation clock". Dev Dyn. 236 (6): 1416–1421. doi: 10.1002/dvdy.21139 . PMID 17420984.