Hydrolysis is any chemical reaction in which a molecule of water breaks one or more chemical bonds. The term is used broadly for substitution, elimination, and solvation reactions in which water is the nucleophile.
Molecular biology is a branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions.

The polymerase chain reaction (PCR) is a method widely used to make millions to billions of copies of a specific DNA sample rapidly, allowing scientists to amplify a very small sample of DNA sufficiently to enable detailed study. PCR was invented in 1983 by American biochemist Kary Mullis at Cetus Corporation. Mullis and biochemist Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993.

The western blot, or western blotting, is a widely used analytical technique in molecular biology and immunogenetics to detect specific proteins in a sample of tissue homogenate or extract. Besides detecting the proteins, this technique is also utilized to visualize, distinguish, and quantify the different proteins in a complicated protein combination.

Reverse transcription polymerase chain reaction (RT-PCR) is a laboratory technique combining reverse transcription of RNA into DNA and amplification of specific DNA targets using polymerase chain reaction (PCR). It is primarily used to measure the amount of a specific RNA. This is achieved by monitoring the amplification reaction using fluorescence, a technique called real-time PCR or quantitative PCR (qPCR). Confusion can arise because some authors use the acronym RT-PCR to denote real-time PCR. In this article, RT-PCR will denote Reverse Transcription PCR. Combined RT-PCR and qPCR are routinely used for analysis of gene expression and quantification of viral RNA in research and clinical settings.

Cytogenetics is essentially a branch of genetics, but is also a part of cell biology/cytology, that is concerned with how the chromosomes relate to cell behaviour, particularly to their behaviour during mitosis and meiosis. Techniques used include karyotyping, analysis of G-banded chromosomes, other cytogenetic banding techniques, as well as molecular cytogenetics such as fluorescence in situ hybridization (FISH) and comparative genomic hybridization (CGH).

Staining is a technique used to enhance contrast in samples, generally at the microscopic level. Stains and dyes are frequently used in histology, in cytology, and in the medical fields of histopathology, hematology, and cytopathology that focus on the study and diagnoses of diseases at the microscopic level. Stains may be used to define biological tissues, cell populations, or organelles within individual cells.
An assay is an investigative (analytic) procedure in laboratory medicine, mining, pharmacology, environmental biology and molecular biology for qualitatively assessing or quantitatively measuring the presence, amount, or functional activity of a target entity. The measured entity is often called the analyte, the measurand, or the target of the assay. The analyte can be a drug, biochemical substance, chemical element or compound, or cell in an organism or organic sample. An assay usually aims to measure an analyte's intensive property and express it in the relevant measurement unit.

Histopathology is the microscopic examination of tissue in order to study the manifestations of disease. Specifically, in clinical medicine, histopathology refers to the examination of a biopsy or surgical specimen by a pathologist, after the specimen has been processed and histological sections have been placed onto glass slides. In contrast, cytopathology examines free cells or tissue micro-fragments.
The first isolation of deoxyribonucleic acid (DNA) was done in 1869 by Friedrich Miescher. DNA extraction is the process of isolating DNA from the cells of an organism isolated from a sample, typically a biological sample such as blood, saliva, or tissue. It involves breaking open the cells, removing proteins and other contaminants, and purifying the DNA so that it is free of other cellular components. The purified DNA can then be used for downstream applications such as PCR, sequencing, or cloning. Currently, it is a routine procedure in molecular biology or forensic analyses.
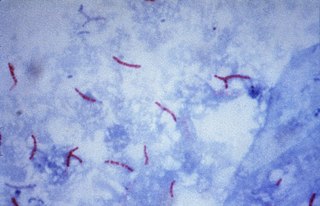
The Ziehl-Neelsen stain, also known as the acid-fast stain, is a bacteriological staining technique used in cytopathology and microbiology to identify acid-fast bacteria under microscopy, particularly members of the Mycobacterium genus. This staining method was initially introduced by Paul Ehrlich (1854–1915) and subsequently modified by the German bacteriologists Franz Ziehl (1859–1926) and Friedrich Neelsen (1854–1898) during the late 19th century.

A fluorescence microscope is an optical microscope that uses fluorescence instead of, or in addition to, scattering, reflection, and attenuation or absorption, to study the properties of organic or inorganic substances. "Fluorescence microscope" refers to any microscope that uses fluorescence to generate an image, whether it is a simple set up like an epifluorescence microscope or a more complicated design such as a confocal microscope, which uses optical sectioning to get better resolution of the fluorescence image.
Torbjörn Oskar Caspersson was a Swedish cytologist and geneticist. He was born in Motala and attended the University of Stockholm, where he studied medicine and biophysics.
Bismarck brown Y also called C.I. 21000 and C.I. Basic Brown 1, is a diazo dye with the idealized formula [(H2N)2C6H3N2]2C6H4. The dye is a mixture of closely related compounds. It was one of the earliest azo dyes, being described in 1863 by German chemist Carl Alexander von Martius. It is used in histology for staining tissues.

Papanicolaou stain is a multichromatic (multicolored) cytological staining technique developed by George Papanicolaou in 1942. The Papanicolaou stain is one of the most widely used stains in cytology, where it is used to aid pathologists in making a diagnosis. Although most notable for its use in the detection of cervical cancer in the Pap test or Pap smear, it is also used to stain non-gynecological specimen preparations from a variety of bodily secretions and from small needle biopsies of organs and tissues. Papanicolaou published three formulations of this stain in 1942, 1954, and 1960.

Hematoxylin and eosin stain is one of the principal tissue stains used in histology. It is the most widely used stain in medical diagnosis and is often the gold standard. For example, when a pathologist looks at a biopsy of a suspected cancer, the histological section is likely to be stained with H&E.

Acridine orange is an organic compound that serves as a nucleic acid-selective fluorescent dye with cationic properties useful for cell cycle determination. Acridine orange is cell-permeable, which allows the dye to interact with DNA by intercalation, or RNA via electrostatic attractions. When bound to DNA, acridine orange is very similar spectrally to an organic compound known as fluorescein. Acridine orange and fluorescein have a maximum excitation at 502nm and 525 nm (green). When acridine orange associates with RNA, the fluorescent dye experiences a maximum excitation shift from 525 nm (green) to 460 nm (blue). The shift in maximum excitation also produces a maximum emission of 650 nm (red). Acridine orange is able to withstand low pH environments, allowing the fluorescent dye to penetrate acidic organelles such as lysosomes and phagolysosomes that are membrane-bound organelles essential for acid hydrolysis or for producing products of phagocytosis of apoptotic cells. Acridine orange is used in epifluorescence microscopy and flow cytometry. The ability to penetrate the cell membranes of acidic organelles and cationic properties of acridine orange allows the dye to differentiate between various types of cells. The shift in maximum excitation and emission wavelengths provides a foundation to predict the wavelength at which the cells will stain.
Virus quantification is counting or calculating the number of virus particles (virions) in a sample to determine the virus concentration. It is used in both research and development (R&D) in academic and commercial laboratories as well as in production situations where the quantity of virus at various steps is an important variable that must be monitored. For example, the production of virus-based vaccines, recombinant proteins using viral vectors, and viral antigens all require virus quantification to continually monitor and/or modify the process in order to optimize product quality and production yields and to respond to ever changing demands and applications. Other examples of specific instances where viruses need to be quantified include clone screening, multiplicity of infection (MOI) optimization, and adaptation of methods to cell culture.
Quantitative Fluorescent in situ hybridization (Q-FISH) is a cytogenetic technique based on the traditional FISH methodology. In Q-FISH, the technique uses labelled synthetic DNA mimics called peptide nucleic acid (PNA) oligonucleotides to quantify target sequences in chromosomal DNA using fluorescent microscopy and analysis software. Q-FISH is most commonly used to study telomere length, which in vertebrates are repetitive hexameric sequences (TTAGGG) located at the distal end of chromosomes. Telomeres are necessary at chromosome ends to prevent DNA-damage responses as well as genome instability. To this day, the Q-FISH method continues to be utilized in the field of telomere research.
Standards for the identification of cell death have changed. Cell death used to be defined and described based on morphology. Now there is a switch in classifying it basing on molecular and genetic definitions. This description is more functional and applies to both in vitro and in vivo, so cell death subroutines are now described by a series of precise, measurable, biochemical features. A set of recommendations for describing the terminology of cell death was proposed by the Nomenclature Committee on Cell Death (NCCD) in 2009, because misusing words and concepts may slow down progress in the area of cell death research.











