Related Research Articles

A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA and RNA. Dictated by specific hydrogen bonding patterns, "Watson–Crick" base pairs allow the DNA helix to maintain a regular helical structure that is subtly dependent on its nucleotide sequence. The complementary nature of this based-paired structure provides a redundant copy of the genetic information encoded within each strand of DNA. The regular structure and data redundancy provided by the DNA double helix make DNA well suited to the storage of genetic information, while base-pairing between DNA and incoming nucleotides provides the mechanism through which DNA polymerase replicates DNA and RNA polymerase transcribes DNA into RNA. Many DNA-binding proteins can recognize specific base-pairing patterns that identify particular regulatory regions of genes.

Nucleotides are organic molecules consisting of a nucleoside and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common nutrients by the liver.
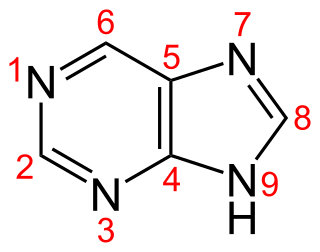
Purine is a heterocyclic aromatic organic compound that consists of two rings fused together. It is water-soluble. Purine also gives its name to the wider class of molecules, purines, which include substituted purines and their tautomers. They are the most widely occurring nitrogen-containing heterocycles in nature.
Pyrimidine is an aromatic heterocyclic organic compound similar to pyridine. One of the three diazines, it has the nitrogen atoms at positions 1 and 3 in the ring. The other diazines are pyrazine and pyridazine. In nucleic acids, three types of nucleobases are pyrimidine derivatives: cytosine (C), thymine (T), and uracil (U).

Nucleobases, also known as nitrogenous bases or often simply bases, are nitrogen-containing biological compounds that form nucleosides, which, in turn, are components of nucleotides, with all of these monomers constituting the basic building blocks of nucleic acids. The ability of nucleobases to form base pairs and to stack one upon another leads directly to long-chain helical structures such as ribonucleic acid (RNA) and deoxyribonucleic acid (DNA).Five nucleobases—adenine (A), cytosine (C), guanine (G), thymine (T), and uracil (U)—are called primary or canonical. They function as the fundamental units of the genetic code, with the bases A, G, C, and T being found in DNA while A, G, C, and U are found in RNA. Thymine and uracil are distinguished by merely the presence or absence of a methyl group on the fifth carbon (C5) of these heterocyclic six-membered rings. In addition, some viruses have aminoadenine (Z) instead of adenine. It differs in having an extra amine group, creating a more stable bond to thymine.
The coding region of a gene, also known as the coding DNA sequence(CDS), is the portion of a gene's DNA or RNA that codes for protein. Studying the length, composition, regulation, splicing, structures, and functions of coding regions compared to non-coding regions over different species and time periods can provide a significant amount of important information regarding gene organization and evolution of prokaryotes and eukaryotes. This can further assist in mapping the human genome and developing gene therapy.

A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create two identical DNA duplexes from a single original DNA duplex. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones. These enzymes catalyze the chemical reaction

In biochemistry, a ribonucleotide is a nucleotide containing ribose as its pentose component. It is considered a molecular precursor of nucleic acids. Nucleotides are the basic building blocks of DNA and RNA. Ribonucleotides themselves are basic monomeric building blocks for RNA. Deoxyribonucleotides, formed by reducing ribonucleotides with the enzyme ribonucleotide reductase (RNR), are essential building blocks for DNA. There are several differences between DNA deoxyribonucleotides and RNA ribonucleotides. Successive nucleotides are linked together via phosphodiester bonds.

Chargaff's rules state that DNA from any species of any organism should have a 1:1 stoichiometric ratio of purine and pyrimidine bases and, more specifically, that the amount of guanine should be equal to cytosine and the amount of adenine should be equal to thymine. This pattern is found in both strands of the DNA. They were discovered by Austrian-born chemist Erwin Chargaff, in the late 1940s.
Satellite DNA consists of very large arrays of tandemly repeating, non-coding DNA. Satellite DNA is the main component of functional centromeres, and form the main structural constituent of heterochromatin.
D-loop replication is a proposed process by which circular DNA like chloroplasts and mitochondria replicate their genetic material. An important component of understanding D-loop replication is that many chloroplasts and mitochondria have a single circular chromosome like bacteria instead of the linear chromosomes found in eukaryotes. However, many chloroplasts and mitochondria have a linear chromosome, and D-loop replication is not important in these organelles. Also, not all circular genomes use D-loop replication as the process of replicating its genome.

Human mitochondrial genetics is the study of the genetics of human mitochondrial DNA. The human mitochondrial genome is the entirety of hereditary information contained in human mitochondria. Mitochondria are small structures in cells that generate energy for the cell to use, and are hence referred to as the "powerhouses" of the cell.
Nuclear DNA (nDNA), or nuclear deoxyribonucleic acid, is the DNA contained within each cell nucleus of a eukaryotic organism. It encodes for the majority of the genome in eukaryotes, with mitochondrial DNA and plastid DNA coding for the rest. It adheres to Mendelian inheritance, with information coming from two parents, one male and one female—rather than matrilineally as in mitochondrial DNA.

In biochemistry and molecular genetics, an AP site, also known as an abasic site, is a location in DNA that has neither a purine nor a pyrimidine base, either spontaneously or due to DNA damage. It has been estimated that under physiological conditions 10,000 apurinic sites and 500 apyrimidinic may be generated in a cell daily.
A polynucleotide molecule is a biopolymer composed of 13 or more nucleotide monomers covalently bonded in a chain. DNA and RNA are examples of polynucleotides with distinct biological function. The prefix poly comes from the ancient Greek πολυς. DNA consists of two chains of polynucleotides, with each chain in the form of a helix.

Nucleic acid metabolism is the process by which nucleic acids are synthesized and degraded. Nucleic acids are the polymers of nucleotides. Nucleotide synthesis is an anabolic mechanism generally involving the chemical reaction of phosphate, pentose sugar, and a nitrogenous base. Destruction of nucleic acid is a catabolic reaction. Additionally, parts of the nucleotides or nucleobases can be salvaged to recreate new nucleotides. Both synthesis and degradation reactions require enzymes to facilitate the event. Defects or deficiencies in these enzymes can lead to a variety of diseases.

Nucleic acid structure refers to the structure of nucleic acids such as DNA and RNA. Chemically speaking, DNA and RNA are very similar. Nucleic acid structure is often divided into four different levels: primary, secondary, tertiary, and quaternary.

Nucleic acid secondary structure is the basepairing interactions within a single nucleic acid polymer or between two polymers. It can be represented as a list of bases which are paired in a nucleic acid molecule. The secondary structures of biological DNAs and RNAs tend to be different: biological DNA mostly exists as fully base paired double helices, while biological RNA is single stranded and often forms complex and intricate base-pairing interactions due to its increased ability to form hydrogen bonds stemming from the extra hydroxyl group in the ribose sugar.
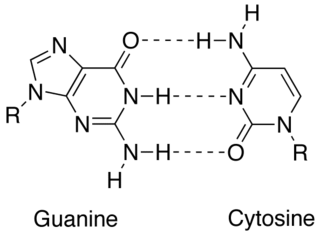
In molecular biology, complementarity describes a relationship between two structures each following the lock-and-key principle. In nature complementarity is the base principle of DNA replication and transcription as it is a property shared between two DNA or RNA sequences, such that when they are aligned antiparallel to each other, the nucleotide bases at each position in the sequences will be complementary, much like looking in the mirror and seeing the reverse of things. This complementary base pairing allows cells to copy information from one generation to another and even find and repair damage to the information stored in the sequences.
xDNA is a size-expanded nucleotide system synthesized from the fusion of a benzene ring and one of the four natural bases: adenine, guanine, cytosine, and thymine. This size expansion produces an 8 letter alphabet which has a larger information density by a factor of 2n compared to natural DNA's 4 letter alphabet. As with normal base-pairing, A pairs with xT, C pairs with xG, G pairs with xC, and T pairs with xA. The double helix is thus 2.4Å wider than a natural double helix. While similar in structure to B-DNA, xDNA has unique absorption, fluorescence, and stacking properties.
References
- ↑ "Heavy Strand - an overview | ScienceDirect Topics". www.sciencedirect.com. Retrieved 2021-12-17.
- ↑ Montoya, J.; Christianson, T.; Levens, D.; Rabinowitz, M.; Attardi, G. (1982-12-01). "Identification of initiation sites for heavy-strand and light-strand transcription in human mitochondrial DNA". Proceedings of the National Academy of Sciences of the United States of America. 79 (23): 7195–7199. doi:10.1073/pnas.79.23.7195. ISSN 0027-8424. PMID 6185947.