Carl Woese was an American microbiologist and biophysicist. Woese is famous for defining the Archaea in 1977 through a pioneering phylogenetic taxonomy of 16S ribosomal RNA, a technique that has revolutionized microbiology. He also originated the RNA world hypothesis in 1967, although not by that name. Woese held the Stanley O. Ikenberry Chair and was professor of microbiology at the University of Illinois Urbana–Champaign.
In biology, a kingdom is the second highest taxonomic rank, just below domain. Kingdoms are divided into smaller groups called phyla.
In biological taxonomy, a domain, also dominion, superkingdom, realm, or empire, is the highest taxonomic rank of all organisms taken together. It was introduced in the three-domain system of taxonomy devised by Carl Woese, Otto Kandler and Mark Wheelis in 1990.
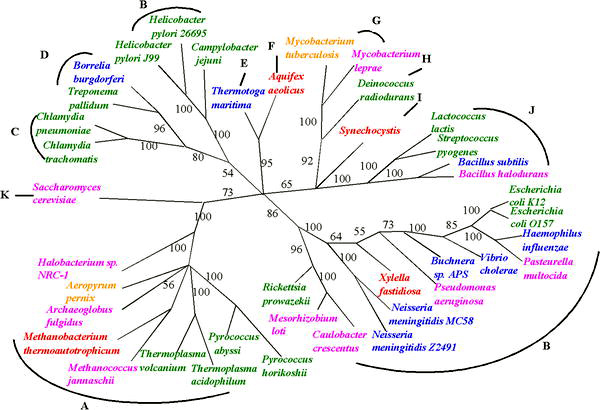
A phylogenetic tree, phylogeny or evolutionary tree is a graphical representation which shows the evolutionary history between a set of species or taxa during a specific time. In other words, it is a branching diagram or a tree showing the evolutionary relationships among various biological species or other entities based upon similarities and differences in their physical or genetic characteristics. In evolutionary biology, all life on Earth is theoretically part of a single phylogenetic tree, indicating common ancestry. Phylogenetics is the study of phylogenetic trees. The main challenge is to find a phylogenetic tree representing optimal evolutionary ancestry between a set of species or taxa. Computational phylogenetics focuses on the algorithms involved in finding optimal phylogenetic tree in the phylogenetic landscape.
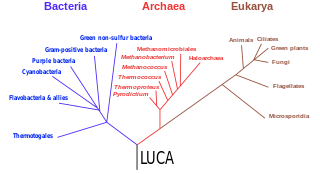
The three-domain system is a taxonomic classification system that groups all cellular life into three domains, namely Archaea, Bacteria and Eukarya, introduced by Carl Woese, Otto Kandler and Mark Wheelis in 1990. The key difference from earlier classifications such as the two-empire system and the five-kingdom classification is the splitting of Archaea from Bacteria as completely different organisms. It has been challenged by the two-domain system that divides organisms into Bacteria and Archaea only, as Eukaryotes are considered as a clade of Archaea.

Horizontal gene transfer (HGT) or lateral gene transfer (LGT) is the movement of genetic material between organisms other than by the ("vertical") transmission of DNA from parent to offspring (reproduction). HGT is an important factor in the evolution of many organisms. HGT is influencing scientific understanding of higher-order evolution while more significantly shifting perspectives on bacterial evolution.

The Thermoproteota are prokaryotes that have been classified as a phylum of the Archaea domain. Initially, the Thermoproteota were thought to be sulfur-dependent extremophiles but recent studies have identified characteristic Thermoproteota environmental rRNA indicating the organisms may be the most abundant archaea in the marine environment. Originally, they were separated from the other archaea based on rRNA sequences; other physiological features, such as lack of histones, have supported this division, although some crenarchaea were found to have histones. Until recently all cultured Thermoproteota had been thermophilic or hyperthermophilic organisms, some of which have the ability to grow at up to 113°C. These organisms stain Gram negative and are morphologically diverse, having rod, cocci, filamentous and oddly-shaped cells.

The last universal common ancestor (LUCA) is the hypothesized common ancestral cell from which the three domains of life, the Bacteria, the Archaea, and the Eukarya originated. It is suggested to have been a "cellular organism that had a lipid bilayer and used DNA, RNA, and protein". The LUCA has also been defined as "a hypothetical organism ancestral to all three domains". The LUCA is the point or stage at which the three domains of life diverged from preexisting forms of life. The nature of this point or stage of divergence remains a topic of research.

Neomura is a proposed clade of biological life composed of the two domains Archaea and Eukaryota, coined by Thomas Cavalier-Smith in 2002. Its name reflects the hypothesis that both archaea and eukaryotes evolved out of the domain Bacteria, and one of the major changes was the replacement of the bacterial peptidoglycan cell walls with other glycoproteins.

Gracilicutes is a clade in bacterial phylogeny.

A prokaryote is a single-cell organism whose cell lacks a nucleus and other membrane-bound organelles. The word prokaryote comes from the Ancient Greek πρό 'before' and κάρυον 'nut, kernel'. In the two-empire system arising from the work of Édouard Chatton, prokaryotes were classified within the empire Prokaryota. But in the three-domain system, based upon molecular analysis, prokaryotes are divided into two domains: Bacteria and Archaea. Organisms with nuclei are placed in a third domain, Eukaryota.

Archaea is a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotes. Archaea were initially classified as bacteria, receiving the name archaebacteria, but this term has fallen out of use.
Evolution of cells refers to the evolutionary origin and subsequent evolutionary development of cells. Cells first emerged at least 3.8 billion years ago approximately 750 million years after Earth was formed.
The eocyte hypothesis in evolutionary biology proposes that the eukaryotes originated from a group of prokaryotes called eocytes. After his team at the University of California, Los Angeles discovered eocytes in 1984, James A. Lake formulated the hypothesis as "eocyte tree" that proposed eukaryotes as part of archaea. Lake hypothesised the tree of life as having only two primary branches: prokaryotes, which include Bacteria and Archaea, and karyotes, that comprise Eukaryotes and eocytes. Parts of this early hypothesis were revived in a newer two-domain system of biological classification which named the primary domains as Archaea and Bacteria.
Microbial phylogenetics is the study of the manner in which various groups of microorganisms are genetically related. This helps to trace their evolution. To study these relationships biologists rely on comparative genomics, as physiology and comparative anatomy are not possible methods.
The Woeseian revolution was the progression of the phylogenetic tree of life concept from two main divisions, known as the Prokarya and Eukarya, into three domains now classified as Bacteria, Archaea, and Eukaryotes. The discovery of the new domain stemmed from the work of biophysicist Carl Woese in 1977 from a principle of evolutionary biology designated as Woese's dogma. It states that the evolution of ribosomal RNA (rRNA) was a necessary precursor to the evolution of modern life forms. Although the three-domain system has been widely accepted, the initial introduction of Woese’s discovery received criticism from the scientific community.
Horizontal or lateral gene transfer is the transmission of portions of genomic DNA between organisms through a process decoupled from vertical inheritance. In the presence of HGT events, different fragments of the genome are the result of different evolutionary histories. This can therefore complicate investigations of the evolutionary relatedness of lineages and species. Also, as HGT can bring into genomes radically different genotypes from distant lineages, or even new genes bearing new functions, it is a major source of phenotypic innovation and a mechanism of niche adaptation. For example, of particular relevance to human health is the lateral transfer of antibiotic resistance and pathogenicity determinants, leading to the emergence of pathogenic lineages.

Darwinian threshold or Darwinian transition is a term introduced by Carl Woese to describe a transition period during the evolution of the first cells when genetic transmission moves from a predominantly horizontal mode to a vertical mode. The process starts when the ancestors of the Last Universal Common Ancestor become refractory to horizontal gene transfer (HGT) and become individual entities with vertical heredity upon which natural selection is effective. After this transition, life is characterized by genealogies that have a modern tree-like phylogeny.

In phylogenetics, reconciliation is an approach to connect the history of two or more coevolving biological entities. The general idea of reconciliation is that a phylogenetic tree representing the evolution of an entity can be drawn within another phylogenetic tree representing an encompassing entity to reveal their interdependence and the evolutionary events that have marked their shared history. The development of reconciliation approaches started in the 1980s, mainly to depict the coevolution of a gene and a genome, and of a host and a symbiont, which can be mutualist, commensalist or parasitic. It has also been used for example to detect horizontal gene transfer, or understand the dynamics of genome evolution.
The first universal common ancestor (FUCA) is a proposed non-cellular entity that is the earliest ancestor of the last universal common ancestor (LUCA) and its descendants, including every modern cell. FUCA would also be the ancestor of ancient sister lineages of LUCA, none of which have modern descendants.