
The genetic code is the set of rules used by living cells to translate information encoded within genetic material into proteins. Translation is accomplished by the ribosome, which links proteinogenic amino acids in an order specified by messenger RNA (mRNA), using transfer RNA (tRNA) molecules to carry amino acids and to read the mRNA three nucleotides at a time. The genetic code is highly similar among all organisms and can be expressed in a simple table with 64 entries.
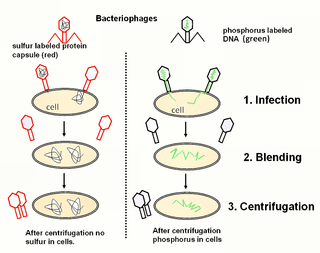
The Hershey–Chase experiments were a series of experiments conducted in 1952 by Alfred Hershey and Martha Chase that helped to confirm that DNA is genetic material.

Ribosomes are macromolecular machines, found within all cells, that perform biological protein synthesis. Ribosomes link amino acids together in the order specified by the codons of messenger RNA molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA molecules and many ribosomal proteins. The ribosomes and associated molecules are also known as the translational apparatus.

In molecular biology, a stop codon is a codon that signals the termination of the translation process of the current protein. Most codons in messenger RNA correspond to the addition of an amino acid to a growing polypeptide chain, which may ultimately become a protein; stop codons signal the termination of this process by binding release factors, which cause the ribosomal subunits to disassociate, releasing the amino acid chain.

Phenylalanine is an essential α-amino acid with the formula C
9H
11NO
2. It can be viewed as a benzyl group substituted for the methyl group of alanine, or a phenyl group in place of a terminal hydrogen of alanine. This essential amino acid is classified as neutral, and nonpolar because of the inert and hydrophobic nature of the benzyl side chain. The L-isomer is used to biochemically form proteins coded for by DNA. Phenylalanine is a precursor for tyrosine, the monoamine neurotransmitters dopamine, norepinephrine (noradrenaline), and epinephrine (adrenaline), and the biological pigment melanin. It is encoded by the messenger RNA codons UUU and UUC.

Transfer RNA is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length, that serves as the physical link between the mRNA and the amino acid sequence of proteins. Transfer RNA (tRNA) does this by carrying an amino acid to the protein-synthesizing machinery of a cell called the ribosome. Complementation of a 3-nucleotide codon in a messenger RNA (mRNA) by a 3-nucleotide anticodon of the tRNA results in protein synthesis based on the mRNA code. As such, tRNAs are a necessary component of translation, the biological synthesis of new proteins in accordance with the genetic code.

The Nirenberg and Matthaei experiment was a scientific experiment performed in May 1961 by Marshall W. Nirenberg and his post-doctoral fellow, J. Heinrich Matthaei, at the National Institutes of Health (NIH). The experiment deciphered the first of the 64 triplet codons in the genetic code by using nucleic acid homopolymers to translate specific amino acids.

The Nirenberg and Leder experiment was a scientific experiment performed in 1964 by Marshall W. Nirenberg and Philip Leder. The experiment elucidated the triplet nature of the genetic code and allowed the remaining ambiguous codons in the genetic code to be deciphered.

Marshall Warren Nirenberg was an American biochemist and geneticist. He shared a Nobel Prize in Physiology or Medicine in 1968 with Har Gobind Khorana and Robert W. Holley for "breaking the genetic code" and describing how it operates in protein synthesis. In the same year, together with Har Gobind Khorana, he was awarded the Louisa Gross Horwitz Prize from Columbia University.
Peter G. Schultz is an American chemist. He is the CEO and Professor of Chemistry at The Scripps Research Institute, the founder and former director of GNF, and the founding director of the California Institute for Biomedical Research (Calibr), established in 2012. In August 2014, Nature Biotechnology ranked Schultz the #1 top translational researcher in 2013.
Xenobiology (XB) is a subfield of synthetic biology, the study of synthesizing and manipulating biological devices and systems. The name "xenobiology" derives from the Greek word xenos, which means "stranger, alien". Xenobiology is a form of biology that is not (yet) familiar to science and is not found in nature. In practice, it describes novel biological systems and biochemistries that differ from the canonical DNA–RNA-20 amino acid system. For example, instead of DNA or RNA, XB explores nucleic acid analogues, termed xeno nucleic acid (XNA) as information carriers. It also focuses on an expanded genetic code and the incorporation of non-proteinogenic amino acids, or “xeno amino acids” into proteins.

A synonymous substitution is the evolutionary substitution of one base for another in an exon of a gene coding for a protein, such that the produced amino acid sequence is not modified. This is possible because the genetic code is "degenerate", meaning that some amino acids are coded for by more than one three-base-pair codon; since some of the codons for a given amino acid differ by just one base pair from others coding for the same amino acid, a mutation that replaces the "normal" base by one of the alternatives will result in incorporation of the same amino acid into the growing polypeptide chain when the gene is translated. Synonymous substitutions and mutations affecting noncoding DNA are often considered silent mutations; however, it is not always the case that the mutation is silent.

Marianne Grunberg-Manago was a Soviet-born French biochemist. Her work helped make possible key discoveries about the nature of the genetic code. Grunberg-Manago was the first woman to lead the International Union of Biochemistry and the 400-year-old French Academy of Sciences.
A cell-free system is an in vitro tool widely used to study biological reactions that happen within cells apart from a full cell system, thus reducing the complex interactions typically found when working in a whole cell. Subcellular fractions can be isolated by ultracentrifugation to provide molecular machinery that can be used in reactions in the absence of many of the other cellular components. Eukaryotic and prokaryotic cell internals have been used for creation of these simplified environments. These systems have enabled cell-free synthetic biology to emerge, providing control over what reaction is being examined, as well as its yield, and lessening the considerations otherwise invoked when working with more sensitive live cells.

An expanded genetic code is an artificially modified genetic code in which one or more specific codons have been re-allocated to encode an amino acid that is not among the 22 common naturally-encoded proteinogenic amino acids.
Cell-free protein synthesis, also known as in vitro protein synthesis or CFPS, is the production of protein using biological machinery in a cell-free system, that is, without the use of living cells. The in vitro protein synthesis environment is not constrained by a cell wall or homeostasis conditions necessary to maintain cell viability. Thus, CFPS enables direct access and control of the translation environment which is advantageous for a number of applications including co-translational solubilisation of membrane proteins, optimisation of protein production, incorporation of non-natural amino acids, selective and site-specific labelling. Due to the open nature of the system, different expression conditions such as pH, redox potentials, temperatures, and chaperones can be screened. Since there is no need to maintain cell viability, toxic proteins can be produced.
The RNA Tie Club was an informal scientific club, meant partly to be humorous, of select scientists who were interested in how proteins were synthesised from genes, specifically the genetic code. It was created by George Gamow upon a suggestion by James Watson in 1954 when the relationship between nucleic acids and amino acids in genetic information was unknown. The club consisted of 20 full members, each representing an amino acid, and four honorary members, representing the four nucleotides. The function of the club members was to think up possible solutions and share with the other members.

A codon table can be used to translate a genetic code into a sequence of amino acids. The standard genetic code is traditionally represented as an RNA codon table, because when proteins are made in a cell by ribosomes, it is messenger RNA (mRNA) that directs protein synthesis. The mRNA sequence is determined by the sequence of genomic DNA. In this context, the standard genetic code is referred to as translation table 1. It can also be represented in a DNA codon table. The DNA codons in such tables occur on the sense DNA strand and are arranged in a 5′-to-3′ direction. Different tables with alternate codons are used depending on the source of the genetic code, such as from a cell nucleus, mitochondrion, plastid, or hydrogenosome.
The history of genetics can be represented on a timeline of events from the earliest work in the 1850s, to the DNA era starting in the 1940s, and the genomics era beginning in the 1970s.
Transcription-translation coupling is a mechanism of gene expression regulation in which synthesis of an mRNA (transcription) is affected by its concurrent decoding (translation). In prokaryotes, mRNAs are translated while they are transcribed. This allows communication between RNA polymerase, the multisubunit enzyme that catalyzes transcription, and the ribosome, which catalyzes translation. Coupling involves both direct physical interactions between RNA polymerase and the ribosome, as well as ribosome-induced changes to the structure and accessibility of the intervening mRNA that affect transcription.













