Related Research Articles

Deoxyribonucleic acid is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of all known organisms and many viruses. DNA and ribonucleic acid (RNA) are nucleic acids. Alongside proteins, lipids and complex carbohydrates (polysaccharides), nucleic acids are one of the four major types of macromolecules that are essential for all known forms of life.

Francis Harry Compton Crick was an English molecular biologist, biophysicist, and neuroscientist. He, James Watson, Rosalind Franklin, and Maurice Wilkins played crucial roles in deciphering the helical structure of the DNA molecule.

Rosalind Elsie Franklin was a British chemist and X-ray crystallographer whose work was central to the understanding of the molecular structures of DNA, RNA, viruses, coal, and graphite. Although her works on coal and viruses were appreciated in her lifetime, Franklin's contributions to the discovery of the structure of DNA were largely unrecognized during her life, for which Franklin has been variously referred to as the "wronged heroine", the "dark lady of DNA", the "forgotten heroine", a "feminist icon", and the "Sylvia Plath of molecular biology".

Susan Lee Lindquist, ForMemRS was an American professor of biology at MIT specializing in molecular biology, particularly the protein folding problem within a family of molecules known as heat-shock proteins, and prions. Lindquist was a member and former director of the Whitehead Institute and was awarded the National Medal of Science in 2010.

A non-coding RNA (ncRNA) is a functional RNA molecule that is not translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important types of non-coding RNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, exRNAs, scaRNAs and the long ncRNAs such as Xist and HOTAIR.

A zinc finger is a small protein structural motif that is characterized by the coordination of one or more zinc ions (Zn2+) which stabilizes the fold. It was originally coined to describe the finger-like appearance of a hypothesized structure from the African clawed frog (Xenopus laevis) transcription factor IIIA. However, it has been found to encompass a wide variety of differing protein structures in eukaryotic cells. Xenopus laevis TFIIIA was originally demonstrated to contain zinc and require the metal for function in 1983, the first such reported zinc requirement for a gene regulatory protein followed soon thereafter by the Krüppel factor in Drosophila. It often appears as a metal-binding domain in multi-domain proteins.

Transfer RNA is an adaptor molecule composed of RNA, typically 76 to 90 nucleotides in length, that serves as the physical link between the mRNA and the amino acid sequence of proteins. Transfer RNA (tRNA) does this by carrying an amino acid to the protein-synthesizing machinery of a cell called the ribosome. Complementation of a 3-nucleotide codon in a messenger RNA (mRNA) by a 3-nucleotide anticodon of the tRNA results in protein synthesis based on the mRNA code. As such, tRNAs are a necessary component of translation, the biological synthesis of new proteins in accordance with the genetic code.

Leslie Eleazer Orgel FRS was a British chemist. He is known for his theories on the origin of life.

The Nirenberg and Matthaei experiment was a scientific experiment performed in May 1961 by Marshall W. Nirenberg and his post-doctoral fellow, J. Heinrich Matthaei, at the National Institutes of Health (NIH). The experiment deciphered the first of the 64 triplet codons in the genetic code by using nucleic acid homopolymers to translate specific amino acids.

The Nirenberg and Leder experiment was a scientific experiment performed in 1964 by Marshall W. Nirenberg and Philip Leder. The experiment elucidated the triplet nature of the genetic code and allowed the remaining ambiguous codons in the genetic code to be deciphered.

Marshall Warren Nirenberg was an American biochemist and geneticist. He shared a Nobel Prize in Physiology or Medicine in 1968 with Har Gobind Khorana and Robert W. Holley for "breaking the genetic code" and describing how it operates in protein synthesis. In the same year, together with Har Gobind Khorana, he was awarded the Louisa Gross Horwitz Prize from Columbia University.
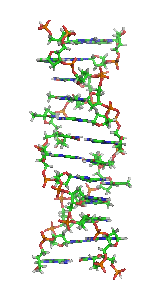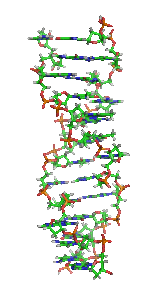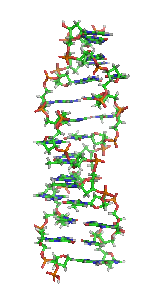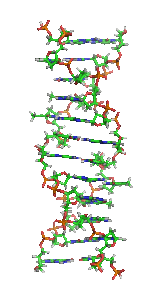
Z-DNA is one of the many possible double helical structures of DNA. It is a left-handed double helical structure in which the helix winds to the left in a zigzag pattern, instead of to the right, like the more common B-DNA form. Z-DNA is thought to be one of three biologically active double-helical structures along with A-DNA and B-DNA.

A Hoogsteen base pair is a variation of base-pairing in nucleic acids such as the A•T pair. In this manner, two nucleobases, one on each strand, can be held together by hydrogen bonds in the major groove. A Hoogsteen base pair applies the N7 position of the purine base and C6 amino group, which bind the Watson–Crick (N3–C4) face of the pyrimidine base.

Venkatraman "Venki" Ramakrishnan is a British-American structural biologist. He shared the 2009 Nobel Prize in Chemistry with Thomas A. Steitz and Ada Yonath for research on the structure and function of ribosomes.
The history of molecular biology begins in the 1930s with the convergence of various, previously distinct biological and physical disciplines: biochemistry, genetics, microbiology, virology and physics. With the hope of understanding life at its most fundamental level, numerous physicists and chemists also took an interest in what would become molecular biology.

The Medical Research Council (MRC) Laboratory of Molecular Biology (LMB) is a research institute in Cambridge, England, involved in the revolution in molecular biology which occurred in the 1950–60s. Since then it has remained a major medical research laboratory at the forefront of scientific discovery, dedicated to improving the understanding of key biological processes at atomic, molecular and cellular levels using multidisciplinary methods, with a focus on using this knowledge to address key issues in human health.
Alexander Rich was an American biologist and biophysicist. He was the William Thompson Sedgwick Professor of Biophysics at MIT and Harvard Medical School. Rich earned an A.B. and an M.D. from Harvard University. He was a post-doc of Linus Pauling. During this time he was a member of the RNA Tie Club, a social and discussion group which attacked the question of how DNA encodes proteins. He has over 600 publications to his name.

The exosome complex is a multi-protein intracellular complex capable of degrading various types of RNA molecules. Exosome complexes are found in both eukaryotic cells and archaea, while in bacteria a simpler complex called the degradosome carries out similar functions.
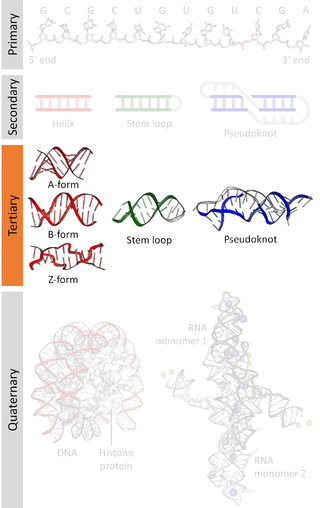
Nucleic acid tertiary structure is the three-dimensional shape of a nucleic acid polymer. RNA and DNA molecules are capable of diverse functions ranging from molecular recognition to catalysis. Such functions require a precise three-dimensional structure. While such structures are diverse and seemingly complex, they are composed of recurring, easily recognizable tertiary structural motifs that serve as molecular building blocks. Some of the most common motifs for RNA and DNA tertiary structure are described below, but this information is based on a limited number of solved structures. Many more tertiary structural motifs will be revealed as new RNA and DNA molecules are structurally characterized.
Éric Westhof is a French biochemist born in Uccle (Belgium) on July 25, 1948. He is a member of the Academie des sciences, head of Education and Training (DEF) and a member of the Board of Directors of the "La Main à la pâte" Foundation. He is professor emeritus of structural biochemistry at the University of Strasbourg at the Institute of Molecular and Cellular Biology.
References
- 1 2 3 Kim SH, Quigley GJ, Suddath FL, et al. (1973). "Three-dimensional structure of yeast phenylalanine transfer RNA: folding of the polynucleotide chain". Science. 179 (4070): 285–8. Bibcode:1973Sci...179..285K. doi:10.1126/science.179.4070.285. PMID 4566654. S2CID 28916938.
- ↑ Tong, L.; Milburn, MV; de Vos, AM; Kim, S.-H. (1989). "Structure of Ras Protein". Science. 245 (244): 244. Bibcode:1989Sci...245..244T. doi:10.1126/science.2665078. PMID 2665078.
- 1 2 3 Suddath FL, Quigley GJ, McPherson A, et al. (1974). "Three-dimensional structure of yeast phenylalanine transfer RNA at 3.0 angstroms resolution". Nature. 248 (443): 20–4. Bibcode:1974Natur.248...20S. doi:10.1038/248020a0. PMID 4594440. S2CID 4207841.
- 1 2 3 Kim SH, Suddath FL, Quigley GJ, et al. (1974). "Three-dimensional tertiary structure of yeast phenylalanine transfer RNA". Science. 185 (4149): 435–40. Bibcode:1974Sci...185..435K. doi:10.1126/science.185.4149.435. PMID 4601792. S2CID 16013012.
- 1 2 Robertus JD, Ladner JE, Finch JT, Rhodes D, et al. (1974). "Structure of yeast phenylalanine tRNA at 3 A resolution". Nature. 250 (467): 546–51. Bibcode:1974Natur.250..546R. doi:10.1038/250546a0. PMID 4602655. S2CID 3051032.
- ↑ Crick's first letter to A. Rich on the accusation, Jul 1974
- ↑ Rich's reply to Crick's first letter, Aug 1974
- ↑ Crick's second letter to Rich, Sept 1974
- ↑ Plexxikon web site