Related Research Articles
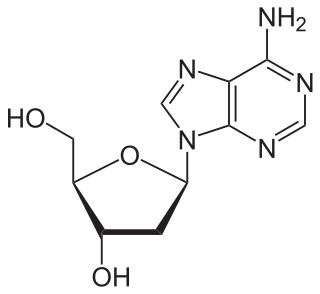
Nucleosides are glycosylamines that can be thought of as nucleotides without a phosphate group. A nucleoside consists simply of a nucleobase and a five-carbon sugar whereas a nucleotide is composed of a nucleobase, a five-carbon sugar, and one or more phosphate groups. In a nucleoside, the anomeric carbon is linked through a glycosidic bond to the N9 of a purine or the N1 of a pyrimidine. Nucleotides are the molecular building blocks of DNA and RNA.
Oligonucleotides are short DNA or RNA molecules, oligomers, that have a wide range of applications in genetic testing, research, and forensics. Commonly made in the laboratory by solid-phase chemical synthesis, these small fragments of nucleic acids can be manufactured as single-stranded molecules with any user-specified sequence, and so are vital for artificial gene synthesis, polymerase chain reaction (PCR), DNA sequencing, molecular cloning and as molecular probes. In nature, oligonucleotides are usually found as small RNA molecules that function in the regulation of gene expression, or are degradation intermediates derived from the breakdown of larger nucleic acid molecules.

A DNA microarray is a collection of microscopic DNA spots attached to a solid surface. Scientists use DNA microarrays to measure the expression levels of large numbers of genes simultaneously or to genotype multiple regions of a genome. Each DNA spot contains picomoles of a specific DNA sequence, known as probes. These can be a short section of a gene or other DNA element that are used to hybridize a cDNA or cRNA sample under high-stringency conditions. Probe-target hybridization is usually detected and quantified by detection of fluorophore-, silver-, or chemiluminescence-labeled targets to determine relative abundance of nucleic acid sequences in the target. The original nucleic acid arrays were macro arrays approximately 9 cm × 12 cm and the first computerized image based analysis was published in 1981. It was invented by Patrick O. Brown. An example of its application is in SNPs arrays for polymorphisms in cardiovascular diseases, cancer, pathogens and GWAS analysis. It is also used for the identification of structural variations and the measurement of gene expression.
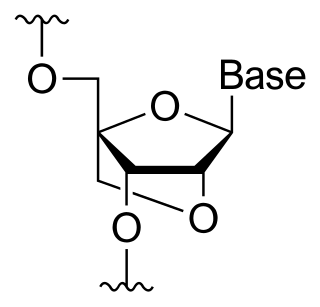
A locked nucleic acid (LNA), also known as bridged nucleic acid (BNA), and often referred to as inaccessible RNA, is a modified RNA nucleotide in which the ribose moiety is modified with an extra bridge connecting the 2' oxygen and 4' carbon. The bridge "locks" the ribose in the 3'-endo (North) conformation, which is often found in the A-form duplexes. This structure provides for increased stability against enzymatic degradation. LNA also offers improved specificity and affinity in base-pairing as a monomer or a constituent of an oligonucleotide. LNA nucleotides can be mixed with DNA or RNA residues in a oligonucleotide.

A protecting group or protective group is introduced into a molecule by chemical modification of a functional group to obtain chemoselectivity in a subsequent chemical reaction. It plays an important role in multistep organic synthesis.
Xenobiology (XB) is a subfield of synthetic biology, the study of synthesizing and manipulating biological devices and systems. The name "xenobiology" derives from the Greek word xenos, which means "stranger, alien". Xenobiology is a form of biology that is not (yet) familiar to science and is not found in nature. In practice, it describes novel biological systems and biochemistries that differ from the canonical DNA–RNA-20 amino acid system. For example, instead of DNA or RNA, XB explores nucleic acid analogues, termed xeno nucleic acid (XNA) as information carriers. It also focuses on an expanded genetic code and the incorporation of non-proteinogenic amino acids, or “xeno amino acids” into proteins.
In chemistry, solid-phase synthesis is a method in which molecules are covalently bound on a solid support material and synthesised step-by-step in a single reaction vessel utilising selective protecting group chemistry. Benefits compared with normal synthesis in a liquid state include:
Deoxyribozymes, also called DNA enzymes, DNAzymes, or catalytic DNA, are DNA oligonucleotides that are capable of performing a specific chemical reaction, often but not always catalytic. This is similar to the action of other biological enzymes, such as proteins or ribozymes . However, in contrast to the abundance of protein enzymes in biological systems and the discovery of biological ribozymes in the 1980s, there is only little evidence for naturally occurring deoxyribozymes. Deoxyribozymes should not be confused with DNA aptamers which are oligonucleotides that selectively bind a target ligand, but do not catalyze a subsequent chemical reaction.
Threose nucleic acid (TNA) is an artificial genetic polymer in which the natural five-carbon ribose sugar found in RNA has been replaced by an unnatural four-carbon threose sugar. Invented by Albert Eschenmoser as part of his quest to explore the chemical etiology of RNA, TNA has become an important synthetic genetic polymer (XNA) due to its ability to efficiently base pair with complementary sequences of DNA and RNA. The main difference between TNA and DNA/RNA is their backbones. DNA and RNA have their phosphate backbones attached to the 5' carbon of the deoxyribose or ribose sugar ring, respectively. TNA, on the other hand, has it's phosphate backbone directly attached to the 3' carbon in the ring, since it does not have a 5' carbon. This modified backbone makes TNA, unlike DNA and RNA, completely refractory to nuclease digestion, making it a promising nucleic acid analog for therapeutic and diagnostic applications.

Glycol nucleic acid (GNA), sometimes also referred to as glycerol nucleic acid, is a nucleic acid similar to DNA or RNA but differing in the composition of its sugar-phosphodiester backbone, using propylene glycol in place of ribose or deoxyribose. GNA is chemically stable but not known to occur naturally. However, due to its simplicity, it might have played a role in the evolution of life.
Purine nucleoside phosphorylase, PNP, PNPase or inosine phosphorylase is an enzyme that in humans is encoded by the NP gene. It catalyzes the chemical reaction
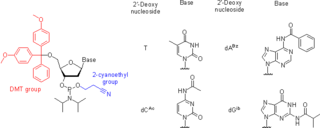
Nucleoside phosphoramidites are derivatives of natural or synthetic nucleosides. They are used to synthesize oligonucleotides, relatively short fragments of nucleic acid and their analogs. Nucleoside phosphoramidites were first introduced in 1981 by Beaucage and Caruthers. To avoid undesired side reactions, reactive hydroxy and exocyclic amino groups present in natural or synthetic nucleosides are appropriately protected. As long as a nucleoside analog contains at least one hydroxy group, the use of the appropriate protecting strategy allows one to convert that to the respective phosphoramidite and to incorporate the latter into synthetic nucleic acids. To be incorporated in the middle of an oligonucleotide chain using phosphoramidite strategy, the nucleoside analog must possess two hydroxy groups or, less often, a hydroxy group and another nucleophilic group (amino or mercapto). Examples include, but are not limited to, alternative nucleotides, LNA, morpholino, nucleosides modified at the 2'-position (OMe, protected NH2, F), nucleosides containing non-canonical bases (hypoxanthine and xanthine contained in natural nucleosides inosine and xanthosine, respectively, tricyclic bases such as G-clamp, etc.) or bases derivatized with a fluorescent group or a linker arm.

Aminoallyl nucleotide is a nucleotide with a modified base containing an allylamine. They are used in post-labeling of nucleic acids by fluorescence detection in microarray. They are reactive with N-Hydroxysuccinimide ester group which helps attach a fluorescent dye to the primary amino group on the nucleotide. These nucleotides are known as 5-(3-aminoallyl)-nucleotides since the aminoallyl group is usually attached to carbon 5 of the pyrimidine ring of uracil or cytosine. The primary amine group in the aminoallyl moiety is aliphatic and thus more reactive compared to the amine groups that are directly attached to the rings (aromatic) of the bases. Common names of aminoallyl nucleosides are initially abbreviated with aa- or AA- to indicate aminoallyl. The 5-carbon sugar is indicated with or without the lowercase "d" indicating deoxyribose if included or ribose if not. Finally the nitrogenous base and number of phosphates are indicated.
Oligonucleotide synthesis is the chemical synthesis of relatively short fragments of nucleic acids with defined chemical structure (sequence). The technique is extremely useful in current laboratory practice because it provides a rapid and inexpensive access to custom-made oligonucleotides of the desired sequence. Whereas enzymes synthesize DNA and RNA only in a 5' to 3' direction, chemical oligonucleotide synthesis does not have this limitation, although it is most often carried out in the opposite, 3' to 5' direction. Currently, the process is implemented as solid-phase synthesis using phosphoramidite method and phosphoramidite building blocks derived from protected 2'-deoxynucleosides, ribonucleosides, or chemically modified nucleosides, e.g. LNA or BNA.

Nucleic acid analogues are compounds which are analogous to naturally occurring RNA and DNA, used in medicine and in molecular biology research. Nucleic acids are chains of nucleotides, which are composed of three parts: a phosphate backbone, a pentose sugar, either ribose or deoxyribose, and one of four nucleobases. An analogue may have any of these altered. Typically the analogue nucleobases confer, among other things, different base pairing and base stacking properties. Examples include universal bases, which can pair with all four canonical bases, and phosphate-sugar backbone analogues such as PNA, which affect the properties of the chain . Nucleic acid analogues are also called Xeno Nucleic Acid and represent one of the main pillars of xenobiology, the design of new-to-nature forms of life based on alternative biochemistries.

Nucleic acid templated chemistry (NATC), or DNA-templated chemistry, is a tool used in the controlled synthesis of chemical compounds. The main advantage of NAT-chemistry (NATC) is that it allows the user to perform the chemical reaction as an intramolecular reaction. Two oligonucleotides. or their analogues, are linked via chemical groups to precursors of chemical compounds. The oligonucleotides recognize specific nucleic acids and are hybridized sterically close to each other. Afterwards, the chemical active groups interact with each other to combine the precursors into a completely new chemical compound. NATC is usually used to perform synthesis of complex compounds without the need to protect chemically active groups during the synthesis.
A bridged nucleic acid (BNA) is a modified RNA nucleotide. They are sometimes also referred to as constrained or inaccessible RNA molecules. BNA monomers can contain a five-membered, six-membered or even a seven-membered bridged structure with a "fixed" C3'-endo sugar puckering. The bridge is synthetically incorporated at the 2', 4'-position of the ribose to afford a 2', 4'-BNA monomer. The monomers can be incorporated into oligonucleotide polymeric structures using standard phosphoramidite chemistry. BNAs are structurally rigid oligo-nucleotides with increased binding affinities and stability.
Thermolabile Protecting Groups (TPGs) are applied in chemical synthesis when mild deprotection conditions are required. Their removal merely consists of increasing temperature, which leads to deprotection of the protected sensitive part of a molecule.

Michal Hocek is a Czech chemist. He is a group leader at the Institute of Organic Chemistry and Biochemistry of the Czech Academy of Sciences and a professor of organic chemistry at Charles University in Prague. He specializes in the chemistry and chemical biology of nucleosides, nucleotides, and nucleic acids.
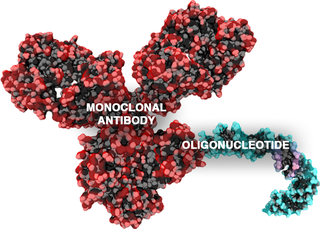
Antibody-oligonucleotide conjugates or AOCs belong to a class of chimeric molecules combining in their structure two important families of biomolecules: monoclonal antibodies and oligonucleotides.
References
- 1 2 Biography Archived 2011-07-11 at the Wayback Machine as a speaker at the Girindus Leadership in Oligonucleotide Symposium, retrieved 2010-09-12.
- ↑ Watts, J.K.; Damha, M.J. (2008). "2′F-Arabinonucleic acids (2′F-ANA) — History, properties, and new frontiers". Can. J. Chem. 86 (7): 641–656. doi:10.1139/V08-049.
- ↑ Damha, M. J.; Wilds, C. J.; Noronha, A.; Brukner, I.; Borkow, G.; Arion, D.; Parniak, M. A. (1998). "Hybrids of RNA and arabinonucleic acids (ANA and 2'F-ANA) are substrates of Ribonuclease H". J. Am. Chem. Soc. 120 (49): 12976–7. doi:10.1021/ja982325.
- ↑ "Archived copy" (PDF). Archived from the original (PDF) on 2016-03-03. Retrieved 2009-11-04.
{{cite web}}: CS1 maint: archived copy as title (link) - ↑ Carriero, S.; Damha, M.J (2003). "Template-Mediated Synthesis of Lariat RNA and DNA". J. Org. Chem. 68 (22): 8328–8338. doi:10.1021/jo035002r. PMID 14575454.
- ↑ Mitra, D.; Damha, M.J. (2007). "A Novel Approach to the Synthesis of DNA and RNA Lariats". J. Org. Chem. 72 (25): 9491–9500. doi:10.1021/jo701418w. PMID 17979285.
- ↑ Lackey, J.G.; Sabatino, D; Damha, M.J (2007). "Solid-Phase Synthesis and On-Column Deprotection of RNA from 2'- (and 3'-) O-Levulinated (Lv) Ribonucleoside Monomers". Org. Lett. 9 (5): 789–792. doi:10.1021/ol0629521. PMID 17279762.
- ↑ Jeremy G. Lackey; Debbie Mitra; Mark M. Somoza; Franco Cerrina; Masad J. Damha (2009). "Acetal Levulinyl Ester (ALE) Groups for 2′-Hydroxyl Protection of Ribonucleosides in the Synthesis of Oligoribonucleotides on Glass and Microarrays". J. Am. Chem. Soc. 131 (24): 8496–8502. doi:10.1021/ja9002074. PMID 19485360.
- ↑ Canadian Society for Chemistry 2007 award winners, Canadian Chemical News, May 1, 2007.
- ↑ Belleu Award Recipients Archived 2010-12-27 at the Wayback Machine , Canadian Society for Chemistry, retrieved 2010-09-12.