Related Research Articles

Streptomyces is the largest genus of Actinomycetota, and the type genus of the family Streptomycetaceae. Over 700 species of Streptomyces bacteria have been described. As with the other Actinomycetota, streptomycetes are gram-positive, and have very large genomes with high GC content. Found predominantly in soil and decaying vegetation, most streptomycetes produce spores, and are noted for their distinct "earthy" odor that results from production of a volatile metabolite, geosmin. Different strains of the same species may colonize very diverse environments.
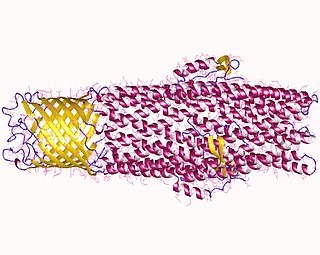
In microbiology, efflux is the moving of a variety of different compounds out of cells, such as antibiotics, heavy metals, organic pollutants, plant-produced compounds, quorum sensing signals, bacterial metabolites and neurotransmitters. All microorganisms, with a few exceptions, have highly conserved DNA sequences in their genome that encode efflux pumps. Efflux pumps actively move substances out of a microorganism, in a process known as active efflux, which is a vital part of xenobiotic metabolism. This active efflux mechanism is responsible for various types of resistance to bacterial pathogens within bacterial species - the most concerning being antibiotic resistance because microorganisms can have adapted efflux pumps to divert toxins out of the cytoplasm and into extracellular media.

Streptomyces griseus is a species of bacteria in the genus Streptomyces commonly found in soil. A few strains have been also reported from deep-sea sediments. It is a Gram-positive bacterium with high GC content. Along with most other streptomycetes, S. griseus strains are well known producers of antibiotics and other such commercially significant secondary metabolites. These strains are known to be producers of 32 different structural types of bioactive compounds. Streptomycin, the first antibiotic ever reported from a bacterium, comes from strains of S. griseus. Recently, the whole genome sequence of one of its strains had been completed.
Sir David Alan Hopwood is a British microbiologist and geneticist.

Actinorhodin is a benzoisochromanequinone dimer polyketide antibiotic produced by Streptomyces coelicolor. The gene cluster responsible for actinorhodin production contains the biosynthetic enzymes and genes responsible for export of the antibiotic. The antibiotic also has the effect of being a pH indicator due to its pH-dependent color change.
Plasmid-mediated resistance is the transfer of antibiotic resistance genes which are carried on plasmids. Plasmids possess mechanisms that ensure their independent replication as well as those that regulate their replication number and guarantee stable inheritance during cell division. By the conjugation process, they can stimulate lateral transfer between bacteria from various genera and kingdoms. Numerous plasmids contain addiction-inducing systems that are typically based on toxin-antitoxin factors and capable of killing daughter cells that don't inherit the plasmid during cell division. Plasmids often carry multiple antibiotic resistance genes, contributing to the spread of multidrug-resistance (MDR). Antibiotic resistance mediated by MDR plasmids severely limits the treatment options for the infections caused by Gram-negative bacteria, especially family Enterobacteriaceae. The global spread of MDR plasmids has been enhanced by selective pressure from antimicrobial medications used in medical facilities and when raising animals for food.
Streptomyces avermitilis is a species of bacteria in the genus Streptomyces. This bacterium was discovered by Satoshi Ōmura in Shizuoka Prefecture, Japan.
Geobacter metallireducens is a gram-negative metal-reducing proteobacterium. It is a strict anaerobe that oxidizes several short-chain fatty acids, alcohols, and monoaromatic compounds with Fe(III) as the sole electron acceptor. It can also use uranium for its growth and convert U(VI) to U(IV).
Streptomyces isolates have yielded the majority of human, animal, and agricultural antibiotics, as well as a number of fundamental chemotherapy medicines. Streptomyces is the largest antibiotic-producing genus of Actinomycetota, producing chemotherapy, antibacterial, antifungal, antiparasitic drugs, and immunosuppressants. Streptomyces isolates are typically initiated with the aerial hyphal formation from the mycelium.
Streptomyces albidoflavus is a bacterium species from the genus of Streptomyces which has been isolated from soil from Poland. Streptomyces albidoflavus produces dibutyl phthalate and streptothricins.
Streptomyces azureus is a bacterium species from the genus of Streptomyces which has isolated from soil. Streptomyces azureus produces the antibiotic thiostrepton.
Streptomyces cacaoi is a bacterium species from the genus of Streptomyces. Streptomyces cacaoi produces polyoxine.
Streptomyces lavendulae is a species of bacteria from the genus Streptomyces. It is isolated from soils globally and is known for its production of medically useful biologically active metabolites. To see a photo of this organism click here.
Streptomyces echinatus is a bacterium species from the genus of Streptomyces which was isolated from soil in Angola. Streptomyces echinatus produces echinomycin, dehydrosinefungin A and aranciamycin.
Streptomyces glaucescens is a bacterium species from the genus of Streptomyces which has been isolated from soil. Streptomyces glaucescens produces tetracenomycin C, tetracenomycin D and tetracenomycin E.
Streptomyces sanglieri is a bacterium species from the genus of Streptomyces which has been isolated from soil from a hay meadow. Streptomyces sanglieri produces the antibiotic lactonamycin Z.
Streptomyces vietnamensis is a bacterium species from the genus of Streptomyces which has been isolated from forest soil in Vietnam.
Streptomyces violaceolatus is a bacterium species from the genus of Streptomyces.
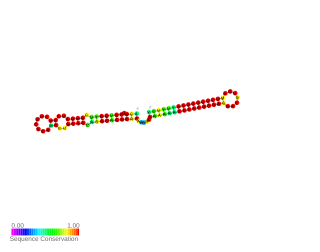
s-SodF RNA is a non-coding RNA (ncRNA) molecule identified in Streptomyces coelicolor. It is produced from sodF mRNA by cleavage of about 90 nucleotides from its 3′UTR. However it does not affect the function of sodF mRNA, but It acts on another mRNA called sodN. s-SodF RNA has a sequence complementary to sodN mRNA from the 5′-end up to the ribosome binding site. It pairs with sodN mRNA, blocks its translation and facilitates sodN mRNA decay. In Streptomyces sodF and sodN genes produce FeSOD and NiSOD superoxide dismutases containing Fe and Ni respectively. Their expression is inversely regulated by nickel-specific Fur-family regulator called Nur. When Ni is present Nur directly represses sodF transcription, and indirectly induces sodN.
Cytochrome P450, family 105, also known as CYP105, is a cytochrome P450 monooxygenase family in bacteria, predominantly found in the phylum Actinomycetota and the order Actinomycetales. The first three genes and subfamilies identified in this family is the herbicide-inducible P-450SU1 and P-450SU2 from Streptomyces griseolus and choP from Streptomyces sp's cholesterol oxidase promoter region.
References
- 1 2 http://royalsociety.org/people/mervyn-bibb/ [ dead link ]
- ↑ Mervyn Bibb publications indexed by Google Scholar
- ↑ Bibb, M; Schottel, J. L.; Cohen, S. N. (1980). "A DNA cloning system for interspecies gene transfer in antibiotic-producing Streptomyces". Nature. 284 (5756): 526–31. Bibcode:1980Natur.284..526B. doi:10.1038/284526a0. PMID 7366721. S2CID 25606114.
- 1 2 Q&A with Professor Mervyn Bibb
- ↑ Bibb, M. J.; Ward, J. M.; Hopwood, D. A. (1978). "Transformation of plasmid DNA into Streptomyces at high frequency". Nature. 274 (5669): 398–400. Bibcode:1978Natur.274..398B. doi:10.1038/274398a0. PMID 672966. S2CID 4221380.
- ↑ Freeman, R. F.; Bibb, M. J.; Hopwood, D. A. (1977). "Chloramphenicol acetylransferase-independent chloramphenicol resistance in Streptomyces coelicolor A3(2)". Journal of General Microbiology. 98 (2): 453–65. doi: 10.1099/00221287-98-2-453 . PMID 856941.
- ↑ Mervyn Bibb's publications indexed by the Scopus bibliographic database. (subscription required)
- ↑ Bibb, Mervyn (1978). Genetic and physical studies of a Streptomyces coelicolor plasmid (PhD thesis). University of East Anglia.
- ↑ Bibb, M. J.; Findlay, P. R.; Johnson, M. W. (1984). "The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences". Gene. 30 (1–3): 157–66. doi:10.1016/0378-1119(84)90116-1. PMID 6096212.
- ↑ Ward, J. M.; Janssen, G. R.; Kieser, T.; Bibb, M. J.; Buttner, M. J.; Bibb, M. J. (1986). "Construction and characterisation of a series of multi-copy promoter-probe plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator". MGG Molecular & General Genetics. 203 (3): 468–478. doi:10.1007/BF00422072. PMID 3018431. S2CID 25943568.
- ↑ Bibb, M. J. (2005). "Regulation of secondary metabolism in streptomycetes". Current Opinion in Microbiology. 8 (2): 208–15. doi:10.1016/j.mib.2005.02.016. PMID 15802254.