
Mycoplasma genitalium is a sexually transmitted, small and pathogenic bacterium that lives on the mucous epithelial cells of the urinary and genital tracts in humans. Medical reports published in 2007 and 2015 state that Mgen is becoming increasingly common. Resistance to multiple antibiotics is becoming prevalent, including to azithromycin, which until recently was the most reliable treatment. The bacteria was first isolated from the urogenital tract of humans in 1981, and was eventually identified as a new species of Mycoplasma in 1983. It can cause negative health effects in men and women. It also increases the risk factor for HIV spread with higher occurrences in those previously treated with the azithromycin antibiotics.

An infection is the invasion of tissues by pathogens, their multiplication, and the reaction of host tissues to the infectious agent and the toxins they produce. An infectious disease, also known as a transmissible disease or communicable disease, is an illness resulting from an infection.

Mycoplasma is a genus of bacteria that, like the other members of the class Mollicutes, lack a cell wall around their cell membranes. Peptidoglycan (murein) is absent. This characteristic makes them naturally resistant to antibiotics that target cell wall synthesis. They can be parasitic or saprotrophic. Several species are pathogenic in humans, including M. pneumoniae, which is an important cause of "walking" pneumonia and other respiratory disorders, and M. genitalium, which is believed to be involved in pelvic inflammatory diseases. Mycoplasma species are among the smallest organisms yet discovered, can survive without oxygen, and come in various shapes. For example, M. genitalium is flask-shaped, while M. pneumoniae is more elongated, many Mycoplasma species are coccoid. Hundreds of Mycoplasma species infect animals.
Mycoplasma hominis is a species of bacteria in the genus Mycoplasma. M. hominis has the ability to penetrate the interior of human cells. Along with ureaplasmas, mycoplasmas are the smallest free-living organisms known.
Mycoplasma pneumoniae is a very small bacterium in the class Mollicutes. It is a human pathogen that causes the disease mycoplasma pneumonia, a form of atypical bacterial pneumonia related to cold agglutinin disease. M. pneumoniae is characterized by the absence of a peptidoglycan cell wall and resulting resistance to many antibacterial agents. The persistence of M. pneumoniae infections even after treatment is associated with its ability to mimic host cell surface composition.
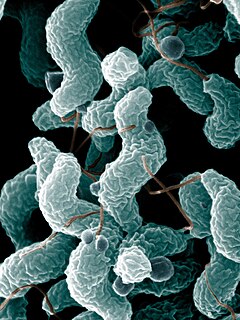
Campylobacter jejuni is one of the most common causes of food poisoning in Europe and in the US. The vast majority of cases occur as isolated events, not as part of recognized outbreaks. Active surveillance through the Foodborne Diseases Active Surveillance Network (FoodNet) indicates that about 20 cases are diagnosed each year for each 100,000 people in the US, while many more cases are undiagnosed or unreported; the CDC estimates a total of 1.5 million infections every year. The European Food Safety Authority reported 246,571 cases in 2018, and estimated approximately nine million cases of human campylobacteriosis per year in the European Union.
Ureaplasma urealyticum is a bacterium belonging to the genus Ureaplasma and the family Mycoplasmataceae in the order Mycoplasmatales. This family consists of the genera Mycoplasma and Ureaplasma. Its type strain is T960. There are two known biovars of this species; T960 and 27. These strains of bacterium are commonly found in the urogenital tracts of human beings, but overgrowth can lead to infections that cause the patient discomfort. Unlike most bacteria, Ureaplasma urealyticum lacks a cell wall making it unique in physiology and medical treatment.
Mollicutes is a class of bacteria distinguished by the absence of a cell wall. The word "Mollicutes" is derived from the Latin mollis, and cutis. Individuals are very small, typically only 0.2–0.3 μm in size and have a very small genome size. They vary in form, although most have sterols that make the cell membrane somewhat more rigid. Many are able to move about through gliding, but members of the genus Spiroplasma are helical and move by twisting. The best-known genus in the Mollicutes is Mycoplasma. Colonies show the typical "fried-egg" appearance.
Mycoplasmataceae is a family of bacteria in the order Mycoplasmatales. This family consists of the genera Mycoplasma and Ureaplasma.

Aeromonas hydrophila is a heterotrophic, Gram-negative, rod-shaped bacterium mainly found in areas with a warm climate. This bacterium can be found in fresh or brackish water. It can survive in aerobic and anaerobic environments, and can digest materials such as gelatin and hemoglobin. A. hydrophila was isolated from humans and animals in the 1950s. It is the best known of the species of Aeromonas. It is resistant to most common antibiotics and cold temperatures and is oxidase- and indole-positive. Aeromonas hydrophila also has a symbiotic relationship as gut flora inside of certain leeches, such as Hirudo medicinalis.
Capnocytophaga canimorsus is a fastidious, slow-growing, Gram-negative rod of the genus Capnocytophaga. It is a commensal bacterium in the normal gingival flora of canine and feline species, but can cause illness in humans. Transmission may occur through bites, licks, or even close proximity with animals. C. canimorsus generally has low virulence in healthy individuals, but has been observed to cause severe, even grave, illness in persons with pre-existing conditions. The pathogenesis of C. canimorsus is still largely unknown, but increased clinical diagnoses have fostered an interest in the bacillus. Treatment with antibiotics is effective in most cases, but the most important yet basic diagnostic tool available to clinicians remains the knowledge of recent exposure to canines or felines.

Bacteria are ubiquitous, mostly free-living organisms often consisting of one biological cell. They constitute a large domain of prokaryotic microorganisms. Typically a few micrometres in length, bacteria were among the first life forms to appear on Earth, and are present in most of its habitats. Bacteria inhabit soil, water, acidic hot springs, radioactive waste, and the deep biosphere of Earth's crust. Bacteria are vital in many stages of the nutrient cycle by recycling nutrients such as the fixation of nitrogen from the atmosphere. The nutrient cycle includes the decomposition of dead bodies; bacteria are responsible for the putrefaction stage in this process. In the biological communities surrounding hydrothermal vents and cold seeps, extremophile bacteria provide the nutrients needed to sustain life by converting dissolved compounds, such as hydrogen sulphide and methane, to energy. Bacteria also live in symbiotic and parasitic relationships with plants and animals. Most bacteria have not been characterised and there are many species that cannot be grown in the laboratory. The study of bacteria is known as bacteriology, a branch of microbiology.
Ureaplasma parvum is a species of Ureaplasma, a genus of bacteria belonging to the family Mycoplasmataceae.
In biology, a pathogen in the oldest and broadest sense, is any organism or agent that can produce disease. A pathogen may also be referred to as an infectious agent, or simply a germ.
Parachlamydia acanthamoebae are bacterium that fall into the category of host-associated microorganisms. This bacterium lives within free-living amoebae that are an intricate part of their reproduction. Originally named Candidatus Parachlamydia acanthamoebae, its current scientific name was introduced shortly after. This species has shown to have over eighty percent 16S rRNA gene sequencing identity with the class Chlamydiia. Parachlamydia acanthamoebae has the same family as the genus Neochlamydia with which it shares many similarities.
Mycoplasma faucium is a species of bacteria in the genus Mycoplasma. This genus of bacteria lacks a cell wall around their cell membrane. Without a cell wall, they are unaffected by many common antibiotics such as penicillin or other beta-lactam antibiotics that target cell wall synthesis. Mycoplasma are the smallest bacterial cells yet discovered, can survive without oxygen and are typically about 0.1 µm in diameter.
Mycoplasma salivarium is a species of bacteria in the genus Mycoplasma. This genus of bacteria lacks a cell wall around their cell membrane. Without a cell wall, they are unaffected by many common antibiotics such as penicillin or other beta-lactam antibiotics that target cell wall synthesis. Mycoplasma are the smallest bacterial cells yet discovered, can survive without oxygen and are typically about 0. 1 μm in diameter. Mycoplasma salivarium is found in the mouths of 97% of the healthy population, and is generally considered to be a commensal organism and part of the normal oral flora.
Mycoplasma alligatoris is a species of bacteria in the genus Mycoplasma. It is classified in the family Mycoplasmataceae, order Mycoplasma, class Mollicutes, phylum Firmicutes and domain Bacteria. Many organisms of the genus Mycoplasma are known pathogens in humans and animal species. Mycoplasma alligatoris is known to elicit a fatal disease with inflammatory characteristics that can cause rapid death of alligators and caimans.
The exact role of Mycoplasma hominis in regards to a number of conditions related to pregnant women and their (unborn) offspring is controversial. This is mainly because many healthy adults have genitourinary colonization with Mycoplasma, published studies on pathogenicity have important design limitations and the organisms are very difficult to detect. The likelihood of colonization with M. hominis appears directly linked to the number of lifetime sexual partners Neonatal colonization does occur, but only through normal vaginal delivery. Caesarean section appears protective against colonization and is much less common. Neonatal colonization is transient.

The uterine microbiome is the commensal, nonpathogenic, bacteria, viruses, yeasts/fungi present in a healthy uterus, amniotic fluid and endometrium and the specific environment which they inhabit. It has been only recently confirmed that the uterus and its tissues are not sterile. Due to improved 16S rRNA gene sequencing techniques, detection of bacteria that are present in low numbers is possible. Using this procedure that allows the detection of bacteria that cannot be cultured outside the body, studies of microbiota present in the uterus are expected to increase.






