Related Research Articles

Histology, also known as microscopic anatomy or microanatomy, is the branch of biology that studies the microscopic anatomy of biological tissues. Histology is the microscopic counterpart to gross anatomy, which looks at larger structures visible without a microscope. Although one may divide microscopic anatomy into organology, the study of organs, histology, the study of tissues, and cytology, the study of cells, modern usage places all of these topics under the field of histology. In medicine, histopathology is the branch of histology that includes the microscopic identification and study of diseased tissue. In the field of paleontology, the term paleohistology refers to the histology of fossil organisms.
The development of the nervous system, or neural development (neurodevelopment), refers to the processes that generate, shape, and reshape the nervous system of animals, from the earliest stages of embryonic development to adulthood. The field of neural development draws on both neuroscience and developmental biology to describe and provide insight into the cellular and molecular mechanisms by which complex nervous systems develop, from nematodes and fruit flies to mammals.

Tomography is imaging by sections or sectioning that uses any kind of penetrating wave. The method is used in radiology, archaeology, biology, atmospheric science, geophysics, oceanography, plasma physics, materials science, cosmochemistry, astrophysics, quantum information, and other areas of science. The word tomography is derived from Ancient Greek τόμος tomos, "slice, section" and γράφω graphō, "to write" or, in this context as well, "to describe." A device used in tomography is called a tomograph, while the image produced is a tomogram.
Brain mapping is a set of neuroscience techniques predicated on the mapping of (biological) quantities or properties onto spatial representations of the brain resulting in maps.
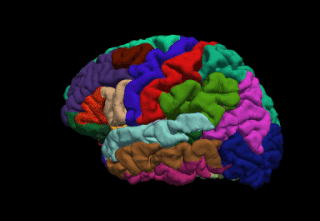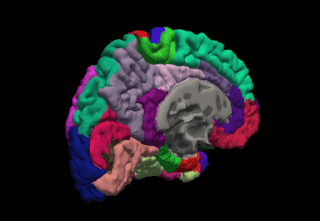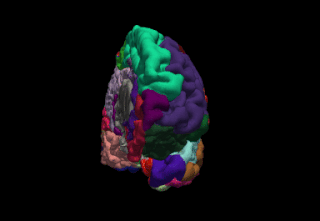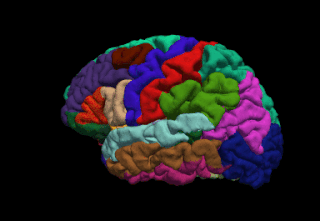
FreeSurfer is a brain imaging software package originally developed by Bruce Fischl, Anders Dale, Martin Sereno, and Doug Greve. Development and maintenance of FreeSurfer is now the primary responsibility of the Laboratory for Computational Neuroimaging at the Athinoula A. Martinos Center for Biomedical Imaging. FreeSurfer contains a set of programs with a common focus of analyzing magnetic resonance imaging (MRI) scans of brain tissue. It is an important tool in functional brain mapping and contains tools to conduct both volume based and surface based analysis. FreeSurfer includes tools for the reconstruction of topologically correct and geometrically accurate models of both the gray/white and pial surfaces, for measuring cortical thickness, surface area and folding, and for computing inter-subject registration based on the pattern of cortical folds.
NeuronStudio was a non-commercial program created at Icahn School of Medicine at Mount Sinai by the Computational Neurobiology and Imaging Center. This program performed automatic tracing and reconstruction of neuron structures from confocal image stacks. The resulting models were then exported to file using standard formats for further processing, modeling, or for statistical analyses. NeuronStudio handled morphologic details on scales spanning local Dendritic spine geometry through complex tree topology to the gross spatial arrangement of multi-neuron networks. Its capability for automated digitization avoided the subjective errors inherent in manual tracing. The program ceased to be supported in 2012 and the project pages were eventually removed from the ISMMS Website. Its documentation and the Windows source code however are still available via the Internet Archive.
Winfried Denk is a German physicist. He built the first two-photon microscope while he was a graduate student in Watt W. Webb's lab at Cornell University, in 1989.
A connectome is a comprehensive map of neural connections in the brain, and may be thought of as its "wiring diagram". An organism's nervous system is made up of neurons which communicate through synapses. A connectome is constructed by tracing the neuron in a nervous system and mapping where neurons are connected through synapses.
Connectomics is the production and study of connectomes: comprehensive maps of connections within an organism's nervous system. More generally, it can be thought of as the study of neuronal wiring diagrams with a focus on how structural connectivity, individual synapses, cellular morphology, and cellular ultrastructure contribute to the make up of a network. The nervous system is a network made of billions of connections and these connections are responsible for our thoughts, emotions, actions, memories, function and dysfunction. Therefore, the study of connectomics aims to advance our understanding of mental health and cognition by understanding how cells in the nervous system are connected and communicate. Because these structures are extremely complex, methods within this field use a high-throughput application of functional and structural neural imaging, most commonly magnetic resonance imaging (MRI), electron microscopy, and histological techniques in order to increase the speed, efficiency, and resolution of these nervous system maps. To date, tens of large scale datasets have been collected spanning the nervous system including the various areas of cortex, cerebellum, the retina, the peripheral nervous system and neuromuscular junctions.
Hyunjune Sebastian Seung is President at Samsung Electronics & Head of Samsung Research and Anthony B. Evnin Professor in the Princeton Neuroscience Institute and Department of Computer Science. Seung has done influential research in both computer science and neuroscience. He has helped pioneer the new field of connectomics, "developing new computational technologies for mapping the connections between neurons," and has been described as the cartographer of the brain.
Serial block-face scanning electron microscopy is a method to generate high resolution three-dimensional images from small samples. The technique was developed for brain tissue, but it is widely applicable for any biological samples. A serial block-face scanning electron microscope consists of an ultramicrotome mounted inside the vacuum chamber of a scanning electron microscope. Samples are prepared by methods similar to that in transmission electron microscopy (TEM), typically by fixing the sample with aldehyde, staining with heavy metals such as osmium and uranium then embedding in an epoxy resin. The surface of the block of resin-embedded sample is imaged by detection of back-scattered electrons. Following imaging the ultramicrotome is used to cut a thin section from the face of the block. After the section is cut, the sample block is raised back to the focal plane and imaged again. This sequence of sample imaging, section cutting and block raising can acquire many thousands of images in perfect alignment in an automated fashion. Practical serial block-face scanning electron microscopy was invented in 2004 by Winfried Denk at the Max-Planck-Institute in Heidelberg and is commercially available from Gatan Inc., Thermo Fisher Scientific (VolumeScope) and ConnectomX.

IMOD is an open-source, cross-platform suite of modeling, display and image processing programs used for 3D reconstruction and modeling of microscopy images with a special emphasis on electron microscopy data. IMOD has been used across a range of scales from macromolecule structures to organelles to whole cells and can also be used for optical sections. IMOD includes tools for image reconstruction, image segmentation, 3D mesh modeling and analysis of 2D and 3D data.
Neuronal tracing, or neuron reconstruction is a technique used in neuroscience to determine the pathway of the neurites or neuronal processes, the axons and dendrites, of a neuron. From a sample preparation point of view, it may refer to some of the following as well as other genetic neuron labeling techniques,
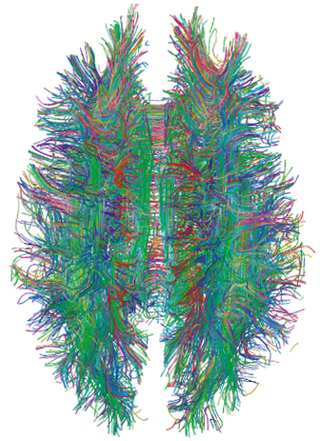
Connectograms are graphical representations of connectomics, the field of study dedicated to mapping and interpreting all of the white matter fiber connections in the human brain. These circular graphs based on diffusion MRI data utilize graph theory to demonstrate the white matter connections and cortical characteristics for single structures, single subjects, or populations.
A Drosophila connectome is a list of neurons in the Drosophila melanogaster nervous system, and the chemical synapses between them. The fly's nervous system consists of the brain plus the ventral nerve cord, and both are known to differ considerably between male and female. Dense connectomes have been completed for the female adult brain, the male nerve cord, and the female larval stage. The available connectomes show only chemical synapses - other forms of inter-neuron communication such as gap junctions or neuromodulators are not represented. Drosophila is the most complex creature with a connectome, which had only been previously obtained for three other simpler organisms, first C. elegans. The connectomes have been obtained by the methods of neural circuit reconstruction, which over the course of many years worked up through various subsets of the fly brain to the almost full connectomes that exist today.
Vaa3D is an Open Source visualization and analysis software suite created mainly by Hanchuan Peng and his team at Janelia Research Campus, HHMI and Allen Institute for Brain Science. The software performs 3D, 4D and 5D rendering and analysis of very large image data sets, especially those generated using various modern microscopy methods, and associated 3D surface objects. This software has been used in several large neuroscience initiatives and a number of applications in other domains. In a recent Nature Methods review article, it has been viewed as one of the leading open-source software suites in the related research fields. In addition, research using this software was awarded the 2012 Cozzarelli Prize from the National Academy of Sciences.
Expansion microscopy (ExM) is a sample preparation tool for biological samples that allows investigators to identify small structures by expanding them using a polymer system. The premise is to introduce a polymer network into cellular or tissue samples, and then physically expand that polymer network using chemical reactions to increase the size of the biological structures. Among other benefits, ExM allows those small structures to be imaged with a wider range of microscopy techniques. It was first proposed in a 2015 article by Fei Chen, Paul W. Tillberg, and Edward Boyden. Current research allows for the expansion of samples up to 16x larger than their initial size. This technique has been found useful in various laboratory settings, such as analyzing biological molecules. ExM allows researchers to use standard equipment in identifying small structures, but requires following of procedures in order to ensure clear results.
Jeff W. Lichtman is an American neuroscientist. He is the Jeremy R. Knowles Professor of Molecular and Cellular Biology and Santiago Ramón y Cajal Professor of Arts and Sciences at Harvard University. He is best known for his pioneering work developing the neuroimaging connectomic technique known as Brainbow.

Carsen Stringer is an American computational neuroscientist and Group Leader at the Howard Hughes Medical Institute Janelia Research Campus. Stringer uses machine learning and deep neural networks to visualize large scale neural recordings and then probe the neural computations that give rise to visual processing in mice. Stringer has also developed several novel software packages that enable cell segmentation and robust analyses of neural recordings and mouse behavior.
Patch-sequencing (patch-seq) is a modification of patch-clamp technique that combines electrophysiological, transcriptomic and morphological characterization of individual neurons. In this approach, the neuron's cytoplasm is collected and processed for RNAseq after electrophysiological recordings are performed on it. The cell is simultaneously filled with a dye that allows for subsequent morphological reconstruction.
References
- 1 2 Chklovskii, Dmitri B; Vitaladevuni, Shiv; Scheffer, Louis K (2010). "Semi-automated reconstruction of neural circuits using electron microscopy". Current Opinion in Neurobiology. 20 (5): 667–75. doi:10.1016/j.conb.2010.08.002. PMID 20833533. S2CID 206950616.
- ↑ Bock, Davi D.; Lee, Wei-Chung Allen; Kerlin, Aaron M.; Andermann, Mark L.; Hood, Greg; Wetzel, Arthur W.; Yurgenson, Sergey; Soucy, Edward R.; et al. (2011). "Network anatomy and in vivo physiology of visual cortical neurons". Nature. 471 (7337): 177–82. Bibcode:2011Natur.471..177B. doi:10.1038/nature09802. PMC 3095821 . PMID 21390124.
- 1 2 White, John G.; Southgate, Eileen; Nichol Thomson, J.; Brenner, Sydney (1986). "The structure of the nervous system of the nematode Caenorhabditis elegans". Philos Trans R Soc Lond B Biol Sci. 314 (1165): 1–340. Bibcode:1986RSPTB.314....1W. doi: 10.1098/rstb.1986.0056 . PMID 22462104.
- ↑ Hayat, M. Arif (2000). Principles and techniques of scanning electron microscopy. Biological applications, fourth edition. Cambridge University Press. ISBN 978-0521632874.
- ↑ Briggman, Kevin L.; Davi D. Bock (2012). "Volume electron microscopy for neuronal circuit reconstruction". Current Opinion in Neurobiology. 22 (1): 154–161. doi:10.1016/j.conb.2011.10.022. PMID 22119321. S2CID 22657332.
- ↑ Saalfeld, Stephan, Albert Cardona, Volker Hartenstein, and Pavel Tomančák (2009). "CATMAID: collaborative annotation toolkit for massive amounts of image data". Bioinformatics. 25 (15): 1984–1986. doi:10.1093/bioinformatics/btp266. PMC 2712332 . PMID 19376822.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ "Large Scale Image Segmentation with Structured Loss based Deep Learning for Connectome Reconstruction". scholar.google.com. Retrieved 2024-02-14.
- ↑ Januszewski, Michał; Kornfeld, Jörgen; Li, Peter H.; Pope, Art; Blakely, Tim; Lindsey, Larry; Maitin-Shepard, Jeremy; Tyka, Mike; Denk, Winfried; Jain, Viren (August 2018). "High-precision automated reconstruction of neurons with flood-filling networks". Nature Methods. 15 (8): 605–610. doi:10.1038/s41592-018-0049-4. ISSN 1548-7105.
- ↑ Chklovskii, Dmitri B., Shiv Vitaladevuni, and Louis K. Scheffer. (2010). "Semi-automated reconstruction of neural circuits using electron microscopy". Current Opinion in Neurobiology. 20 (5): 667–675. doi:10.1016/j.conb.2010.08.002. PMID 20833533. S2CID 206950616.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ↑ Jason Pipkin (Oct 8, 2020). "Connectomes: Mapping the mind of a fly". Elife Sciences.
- ↑ Shapson-Coe, Alexander; Januszewski, Michał; Berger, Daniel R.; Pope, Art; Wu, Yuelong; Blakely, Tim; Schalek, Richard L.; Li, Peter H.; Wang, Shuohong (2021-11-25), A connectomic study of a petascale fragment of human cerebral cortex, doi:10.1101/2021.05.29.446289 , retrieved 2024-02-14
- ↑ Loomba, Sahil; Straehle, Jakob; Gangadharan, Vijayan; Heike, Natalie; Khalifa, Abdelrahman; Motta, Alessandro; Ju, Niansheng; Sievers, Meike; Gempt, Jens; Meyer, Hanno S.; Helmstaedter, Moritz (2022-07-08). "Connectomic comparison of mouse and human cortex". Science. 377 (6602). doi:10.1126/science.abo0924. ISSN 0036-8075.
- ↑ "Analysis tools for connectomics". Howard Hughes Medical Institute.
- ↑ Bargmann, Cornelia I. (2012). "Beyond the connectome: how neuromodulators shape neural circuits". BioEssays. 34 (6): 458–465. doi: 10.1002/bies.201100185 . PMID 22396302.