Related Research Articles
In information science, an ontology encompasses a representation, formal naming, and definitions of the categories, properties, and relations between the concepts, data, or entities that pertain to one, many, or all domains of discourse. More simply, an ontology is a way of showing the properties of a subject area and how they are related, by defining a set of terms and relational expressions that represent the entities in that subject area. The field which studies ontologies so conceived is sometimes referred to as applied ontology.
The Web Ontology Language (OWL) is a family of knowledge representation languages for authoring ontologies. Ontologies are a formal way to describe taxonomies and classification networks, essentially defining the structure of knowledge for various domains: the nouns representing classes of objects and the verbs representing relations between the objects.
The Gene Ontology (GO) is a major bioinformatics initiative to unify the representation of gene and gene product attributes across all species. More specifically, the project aims to: 1) maintain and develop its controlled vocabulary of gene and gene product attributes; 2) annotate genes and gene products, and assimilate and disseminate annotation data; and 3) provide tools for easy access to all aspects of the data provided by the project, and to enable functional interpretation of experimental data using the GO, for example via enrichment analysis. GO is part of a larger classification effort, the Open Biomedical Ontologies, being one of the Initial Candidate Members of the OBO Foundry.

Barry Smith is an academic working in the fields of ontology and biomedical informatics. Smith is the author of more than 700 scientific publications, including 15 authored or edited books, and he is one of the most widely cited living philosophers.
In information science, an upper ontology is an ontology that consists of very general terms that are common across all domains. An important function of an upper ontology is to support broad semantic interoperability among a large number of domain-specific ontologies by providing a common starting point for the formulation of definitions. Terms in the domain ontology are ranked under the terms in the upper ontology, e.g., the upper ontology classes are superclasses or supersets of all the classes in the domain ontologies.
The Open Biological and Biomedical Ontologies (OBO) Foundry is a group of people dedicated to build and maintain ontologies related to the life sciences. The OBO Foundry establishes a set of principles for ontology development for creating a suite of interoperable reference ontologies in the biomedical domain. Currently, there are more than a hundred ontologies that follow the OBO Foundry principles.
The Functional GEnomics Data Society (FGED) was a non-profit, volunteer-run international organization of biologists, computer scientists, and data analysts that aims to facilitate biological and biomedical discovery through data integration. The approach of FGED was to promote the sharing of basic research data generated primarily via high-throughput technologies that generate large data sets within the domain of functional genomics.
Integrative bioinformatics is a discipline of bioinformatics that focuses on problems of data integration for the life sciences.
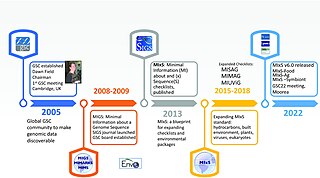
The Genomic Standards Consortium (GSC) is an initiative working towards richer descriptions of our collection of genomes, metagenomes and marker genes. Established in September 2005, this international community includes representatives from a range of major sequencing and bioinformatics centres and research institutions. The goal of the GSC is to promote mechanisms for standardizing the description of (meta)genomes, including the exchange and integration of (meta)genomic data. The number and pace of genomic and metagenomic sequencing projects will only increase as the use of ultra-high-throughput methods becomes common place and standards are vital to scientific progress and data sharing.
Basic Formal Ontology (BFO) is a top-level ontology developed by Barry Smith and his associates for the purposes of promoting interoperability among domain ontologies built in its terms through a process of downward population. A guide to building BFO-conformant domain ontologies was published by MIT Press in 2015.

In computer science, information science and systems engineering, ontology engineering is a field which studies the methods and methodologies for building ontologies, which encompasses a representation, formal naming and definition of the categories, properties and relations between the concepts, data and entities of a given domain of interest. In a broader sense, this field also includes a knowledge construction of the domain using formal ontology representations such as OWL/RDF. A large-scale representation of abstract concepts such as actions, time, physical objects and beliefs would be an example of ontological engineering. Ontology engineering is one of the areas of applied ontology, and can be seen as an application of philosophical ontology. Core ideas and objectives of ontology engineering are also central in conceptual modeling.
The Disease Ontology (DO) is a formal ontology of human disease. The Disease Ontology project is hosted at the Institute for Genome Sciences at the University of Maryland School of Medicine.

Gene set enrichment analysis (GSEA) (also called functional enrichment analysis or pathway enrichment analysis) is a method to identify classes of genes or proteins that are over-represented in a large set of genes or proteins, and may have an association with different phenotypes (e.g. different organism growth patterns or diseases). The method uses statistical approaches to identify significantly enriched or depleted groups of genes. Transcriptomics technologies and proteomics results often identify thousands of genes, which are used for the analysis.

Multiomics, multi-omics, integrative omics, "panomics" or "pan-omics" is a biological analysis approach in which the data sets are multiple "omes", such as the genome, proteome, transcriptome, epigenome, metabolome, and microbiome ; in other words, the use of multiple omics technologies to study life in a concerted way. By combining these "omes", scientists can analyze complex biological big data to find novel associations between biological entities, pinpoint relevant biomarkers and build elaborate markers of disease and physiology. In doing so, multiomics integrates diverse omics data to find a coherently matching geno-pheno-envirotype relationship or association. The OmicTools service lists more than 99 softwares related to multiomic data analysis, as well as more than 99 databases on the topic.
Model organism databases (MODs) are biological databases, or knowledgebases, dedicated to the provision of in-depth biological data for intensively studied model organisms. MODs allow researchers to easily find background information on large sets of genes, plan experiments efficiently, combine their data with existing knowledge, and construct novel hypotheses. They allow users to analyse results and interpret datasets, and the data they generate are increasingly used to describe less well studied species. Where possible, MODs share common approaches to collect and represent biological information. For example, all MODs use the Gene Ontology (GO) to describe functions, processes and cellular locations of specific gene products. Projects also exist to enable software sharing for curation, visualization and querying between different MODs. Organismal diversity and varying user requirements however mean that MODs are often required to customize capture, display, and provision of data.
Minimum information standards are sets of guidelines and formats for reporting data derived by specific high-throughput methods. Their purpose is to ensure the data generated by these methods can be easily verified, analysed and interpreted by the wider scientific community. Ultimately, they facilitate the transfer of data from journal articles into databases in a form that enables data to be mined across multiple data sets. Minimal information standards are available for a vast variety of experiment types including microarray (MIAME), RNAseq (MINSEQE), metabolomics (MSI) and proteomics (MIAPE).
Judith Anne Blake is a computational biologist at the Jackson Laboratory and Professor of Mammalian Genetics.
Biocuration is the field of life sciences dedicated to organizing biomedical data, information and knowledge into structured formats, such as spreadsheets, tables and knowledge graphs. The biocuration of biomedical knowledge is made possible by the cooperative work of biocurators, software developers and bioinformaticians and is at the base of the work of biological databases.

Susanna-Assunta Sansone is a British-Italian data scientist who is professor of data readiness at the University of Oxford where she leads the data readiness group and serves as associate director of the Oxford e-Research Centre. Her research investigates techniques for improving the interoperability, reproducibility and integrity of data.
References
- ↑ Bandrowski A, Brinkman R, Brochhausen M, Brush MH, Bug B, Chibucos MC, et al. (29 April 2016). Xue Y (ed.). "The Ontology for Biomedical Investigations". PLOS ONE. 11 (4): e0154556. Bibcode:2016PLoSO..1154556B. doi: 10.1371/journal.pone.0154556 . PMC 4851331 . PMID 27128319.
- ↑ Ontology for Biomedical Investigations (OBI) | Home
- ↑ "Basic Formal Ontology (BFO)". Institute for Formal Ontology and Medical Information Science. Universität des Saarlandes.
- ↑ Whetzel, PL et al. Development of FuGO: an ontology for functional genomics investigations. OMICS 10, 199–204