Related Research Articles

A mitochondrion is an organelle found in the cells of most eukaryotes, such as animals, plants and fungi. Mitochondria have a double membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is used throughout the cell as a source of chemical energy. They were discovered by Albert von Kölliker in 1857 in the voluntary muscles of insects. The term mitochondrion was coined by Carl Benda in 1898. The mitochondrion is popularly nicknamed the "powerhouse of the cell", a phrase coined by Philip Siekevitz in a 1957 article of the same name.
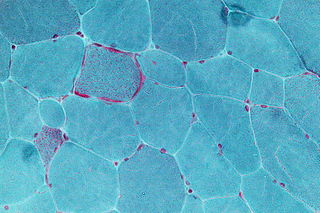
Mitochondrial disease is a group of disorders caused by mitochondrial dysfunction. Mitochondria are the organelles that generate energy for the cell and are found in every cell of the human body except red blood cells. They convert the energy of food molecules into the ATP that powers most cell functions.

Homoplasmy is a term used in genetics to describe a eukaryotic cell whose copies of mitochondrial DNA are all identical. In normal and healthy tissues, all cells are homoplasmic. Homoplasmic mitochondrial DNA copies may be normal or mutated; however, most mutations are heteroplasmic. It has been discovered, though, that homoplasmic mitochondrial DNA mutations may be found in human tumors.

Human mitochondrial genetics is the study of the genetics of human mitochondrial DNA. The human mitochondrial genome is the entirety of hereditary information contained in human mitochondria. Mitochondria are small structures in cells that generate energy for the cell to use, and are hence referred to as the "powerhouses" of the cell.

Leber's hereditary optic neuropathy (LHON) is a mitochondrially inherited degeneration of retinal ganglion cells (RGCs) and their axons that leads to an acute or subacute loss of central vision; it predominantly affects young adult males. LHON is transmitted only through the mother, as it is primarily due to mutations in the mitochondrial genome, and only the egg contributes mitochondria to the embryo. Men cannot pass on the disease to their offspring. LHON is usually due to one of three pathogenic mitochondrial DNA (mtDNA) point mutations. These mutations are at nucleotide positions 11778 G to A, 3460 G to A and 14484 T to C, respectively in the ND4, ND1 and ND6 subunit genes of complex I of the oxidative phosphorylation chain in mitochondria.
Dominant optic atrophy (DOA), or autosomal dominant optic atrophy (ADOA), (Kjer's type) is an autosomally inherited disease that affects the optic nerves, causing reduced visual acuity and blindness beginning in childhood. However, the disease can seem to re-present a second time with further vision loss due to the early onset of presbyopia symptoms (i.e., difficulty in viewing objects up close). DOA is characterized as affecting neurons called retinal ganglion cells (RGCs). This condition is due to mitochondrial dysfunction mediating the death of optic nerve fibers. The RGCs axons form the optic nerve. Therefore, the disease can be considered of the central nervous system. Dominant optic atrophy was first described clinically by Batten in 1896 and named Kjer’s optic neuropathy in 1959 after Danish ophthalmologist Poul Kjer, who studied 19 families with the disease. Although dominant optic atrophy is the most common autosomally inherited optic neuropathy (i.e., disease of the optic nerves), it is often misdiagnosed.
Allotopic expression (AE) refers to expression of genes in the cell nucleus that normally are expressed only from the mitochondrial genome. Biomedically engineered AE has been suggested as a possible future tool in gene therapy of certain mitochondria-related diseases, however this view is controversial. While this type of expression has been successfully carried out in yeast, the results in mammals have been conflicting.

Neuropathy, ataxia, and retinitis pigmentosa, also known as NARP syndrome, is a rare disease with mitochondrial inheritance that causes a variety of signs and symptoms chiefly affecting the nervous system Beginning in childhood or early adulthood, most people with NARP experience numbness, tingling, or pain in the arms and legs ; muscle weakness; and problems with balance and coordination (ataxia). Many affected individuals also have vision loss caused by changes in the light-sensitive tissue that lines the back of the eye. In some cases, the vision loss results from a condition called retinitis pigmentosa. This eye disease causes the light-sensing cells of the retina gradually to deteriorate.

Mitofusin-2 is a protein that in humans is encoded by the MFN2 gene. Mitofusins are GTPases embedded in the outer membrane of the mitochondria. In mammals MFN1 and MFN2 are essential for mitochondrial fusion. In addition to the mitofusins, OPA1 regulates inner mitochondrial membrane fusion, and DRP1 is responsible for mitochondrial fission.

MT-ND6 is a gene of the mitochondrial genome coding for the NADH-ubiquinone oxidoreductase chain 6 protein (ND6). The ND6 protein is a subunit of NADH dehydrogenase (ubiquinone), which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain. Variations in the human MT-ND6 gene are associated with Leigh's syndrome, Leber's hereditary optic neuropathy (LHON) and dystonia.

MT-ND4 is a gene of the mitochondrial genome coding for the NADH-ubiquinone oxidoreductase chain 4 (ND4) protein. The ND4 protein is a subunit of NADH dehydrogenase (ubiquinone), which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain. Variations in the MT-ND4 gene are associated with age-related macular degeneration (AMD), Leber's hereditary optic neuropathy (LHON), mesial temporal lobe epilepsy (MTLE) and cystic fibrosis.

MT-ND2 is a gene of the mitochondrial genome coding for the NADH dehydrogenase 2 (ND2) protein. The ND2 protein is a subunit of NADH dehydrogenase (ubiquinone), which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain. Variants of human MT-ND2 are associated with mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS), Leigh's syndrome (LS), Leber's hereditary optic neuropathy (LHON) and increases in adult BMI.

MT-ND4L is a gene of the mitochondrial genome coding for the NADH-ubiquinone oxidoreductase chain 4L (ND4L) protein. The ND4L protein is a subunit of NADH dehydrogenase (ubiquinone), which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain. Variants of human MT-ND4L are associated with increased BMI in adults and Leber's Hereditary Optic Neuropathy (LHON).

MT-ND5 is a gene of the mitochondrial genome coding for the NADH-ubiquinone oxidoreductase chain 5 protein (ND5). The ND5 protein is a subunit of NADH dehydrogenase (ubiquinone), which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain. Variations in human MT-ND5 are associated with mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS) as well as some symptoms of Leigh's syndrome and Leber's hereditary optic neuropathy (LHON).

MT-ND1 is a gene of the mitochondrial genome coding for the NADH-ubiquinone oxidoreductase chain 1 (ND1) protein. The ND1 protein is a subunit of NADH dehydrogenase, which is located in the mitochondrial inner membrane and is the largest of the five complexes of the electron transport chain. Variants of the human MT-ND1 gene are associated with mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS), Leigh's syndrome (LS), Leber's hereditary optic neuropathy (LHON) and increases in adult BMI.

Cytochrome c oxidase I (COX1) also known as mitochondrially encoded cytochrome c oxidase I (MT-CO1) is a protein that is encoded by the MT-CO1 gene in eukaryotes. The gene is also called COX1, CO1, or COI. Cytochrome c oxidase I is the main subunit of the cytochrome c oxidase complex. In humans, mutations in MT-CO1 have been associated with Leber's hereditary optic neuropathy (LHON), acquired idiopathic sideroblastic anemia, Complex IV deficiency, colorectal cancer, sensorineural deafness, and recurrent myoglobinuria.

Cytochrome c oxidase subunit III (COX3) is an enzyme that in humans is encoded by the MT-CO3 gene. It is one of main transmembrane subunits of cytochrome c oxidase. It is also one of the three mitochondrial DNA (mtDNA) encoded subunits of respiratory complex IV. Variants of it have been associated with isolated myopathy, severe encephalomyopathy, Leber hereditary optic neuropathy, mitochondrial complex IV deficiency, and recurrent myoglobinuria.
Mitochondrial optic neuropathies are a heterogenous group of disorders that present with visual disturbances resultant from mitochondrial dysfunction within the anatomy of the Retinal Ganglion Cells (RGC), optic nerve, optic chiasm, and optic tract. These disturbances are multifactorial, their aetiology consisting of metabolic and/or structural damage as a consequence of genetic mutations, environmental stressors, or both. The three most common neuro-ophthalmic abnormalities seen in mitochondrial disorders are bilateral optic neuropathy, ophthalmoplegia with ptosis, and pigmentary retinopathy.

In biology, mother's curse is an evolutionary effect that males inherit deleterious mitochondrial genome (mtDNA) mutations from their mother, while those mutations are beneficial, neutral or less deleterious to females.

Alfredo Arrigo Sadun is an American ophthalmologist, academic, author and researcher. He holds the Flora L. Thornton Endowed Chair at Doheny Eye Centers-UCLA and is Vice-Chair of Ophthalmology at UCLA.
References
- 1 2 Khan, Shaharyar M.; Bennett, James P. (1 August 2004). "Development of Mitochondrial Gene Replacement Therapy" (PDF). Journal of Bioenergetics and Biomembranes. 36 (4): 387–393. doi:10.1023/B:JOBB.0000041773.20072.9e. PMID 15377877. S2CID 25674350.
- ↑ Aravintha Siva, M.; Mahalakshmi, R.; Bhakta-Guha, Dipita; Guha, Gunjan (1 May 2019). "Gene therapy for the mitochondrial genome: Purging mutations, pacifying ailments". Mitochondrion. 46: 195–208. doi:10.1016/j.mito.2018.06.002. PMID 29890303. S2CID 48356579.
- ↑ Khan, Shaharyar M.; Bennett Jr., James P. (August 2004). "Development of Mitochondrial Gene Replacement Therapy". Journal of Bioenergetics and Biomembranes. 36 (4): 387–393. doi:10.1023/b:jobb.0000041773.20072.9e. ISSN 0145-479X. PMID 15377877. S2CID 25674350.
- ↑ Keeney, Paula M.; Quigley, Caitlin K.; Dunham, Lisa D.; Papageorge, Christina M.; Iyer, Shilpa; Thomas, Ravindar R.; Schwarz, Kathleen M.; Trimmer, Patricia A.; Khan, Shaharyar M.; Portell, Francisco R.; Bergquist, Kristen E.; Bennett, James P. (2009). "Mitochondrial Gene Therapy Augments Mitochondrial Physiology in a Parkinson's Disease Cell Model". Human Gene Therapy. 20 (8): 897–907. doi:10.1089/hum.2009.023. PMC 2829286 . PMID 19374590.
- ↑ Iyer, Shilpa (March 2013). "Novel Therapeutic Approaches for Leber's Hereditary Optic Neuropathy". Discovery Medicine. 15 (82): 141–149. ISSN 1539-6509. PMC 5652312 . PMID 23545042.
- ↑ Rao, Raj R.; Iyer, Shilpa (2015). "Stem Cells, Neural Progenitors, and Engineered Stem Cells". Neuronal Cell Death. Methods in Molecular Biology. Vol. 1254. pp. 255–267. doi:10.1007/978-1-4939-2152-2_19. ISBN 978-1-4939-2151-5. PMC 5642280 . PMID 25431071.
- ↑ Kolata, Gina (10 July 2018). "Dying Organs Restored to Life in Novel Experiments". New York Times . Retrieved 11 July 2018.