An oncogene is a gene that has the potential to cause cancer. In tumor cells, these genes are often mutated, or expressed at high levels.

Desmoplastic small-round-cell tumor (DSRCT) is an aggressive and rare cancer that primarily occurs as masses in the abdomen. Other areas affected may include the lymph nodes, the lining of the abdomen, diaphragm, spleen, liver, chest wall, skull, spinal cord, large intestine, small intestine, bladder, brain, lungs, testicles, ovaries, and the pelvis. Reported sites of metastatic spread include the liver, lungs, lymph nodes, brain, skull, and bones. It is characterized by the EWS-WT1 fusion protein.

The PAX3 gene encodes a member of the paired box or PAX family of transcription factors. The PAX family consists of nine human (PAX1-PAX9) and nine mouse (Pax1-Pax9) members arranged into four subfamilies. Human PAX3 and mouse Pax3 are present in a subfamily along with the highly homologous human PAX7 and mouse Pax7 genes. The human PAX3 gene is located in the 2q36.1 chromosomal region, and contains 10 exons within a 100 kb region.

Ewing sarcoma is a type of pediatric cancer that forms in bone or soft tissue. Symptoms may include swelling and pain at the site of the tumor, fever, and a bone fracture. The most common areas where it begins are the legs, pelvis, and chest wall. In about 25% of cases, the cancer has already spread to other parts of the body at the time of diagnosis. Complications may include a pleural effusion or paraplegia.

Friend leukemia integration 1 transcription factor (FLI1), also known as transcription factor ERGB, is a protein that in humans is encoded by the FLI1 gene, which is a proto-oncogene.

Cyclic AMP-dependent transcription factor ATF-1 is a protein that in humans is encoded by the ATF1 gene.

ETS translocation variant 4 (ETV4), also known as polyoma enhancer activator 3 (PEA3), is a member of the PEA3 subfamily of Ets transcription factors.

The human DEKgene encodes the DEK protein.

ERG is an oncogene. ERG is a member of the ETS family of transcription factors. The ERG gene encodes for a protein, also called ERG, that functions as a transcriptional regulator. Genes in the ETS family regulate embryonic development, cell proliferation, differentiation, angiogenesis, inflammation, and apoptosis.

RNA-binding protein FUS/TLS, also known as heterogeneous nuclear ribonucleoprotein P2 is a protein that in humans is encoded by the FUS gene.

Protein SSX2 is a protein that in humans is encoded by the SSX2 gene.

TATA-binding protein-associated factor 2N is a protein that in humans is encoded by the TAF15 gene.

Ubiquitin carboxyl-terminal hydrolase 6 (USB6), also termed TRE17 and Tre-2, is a deubiquitinating enzyme that in humans is encoded by the hominid USP6 gene located at band 13.2 on the short arm of chromosome 17. Deubiquitinating enzymes (DUBs) are enzymes that act within cells to remove ubiquitins from various functionally important proteins. Ubiquitin enzymes add ubiquitin to these proteins and thereby regulate their cellular location, alter their activity, and/or promote their degradation. By deubiquitinating these proteins, DUBs counter the effects of the ubiquinating enzymes and contribute to regulating the actions of the targeted proteins. In normal adult tissues, USP6 is highly expressed in testicle tissue, modestly expressed in ovarian tissue, and absent or minimally expressed in other tissues. It is also highly expressed in fetal brain tissue. The specific functions of USP6 are poorly defined primarily because its presence is restricted to primates: there are no available animal models to determine the effects of its deletion, although some studies suggest that UPSP6 contributes to normal brain development. In all events, USP6 has gained wide interest because of its abnormally increased expression by the neoplastic cells in various tumors derived from mesenchymal tissue.
In the field of molecular biology, the ETSfamily is one of the largest families of transcription factors and is unique to animals. There are 29 genes in humans, 28 in the mouse, 10 in Caenorhabditis elegans and 9 in Drosophila. The founding member of this family was identified as a gene transduced by the leukemia virus, E26. The members of the family have been implicated in the development of different tissues as well as cancer progression.
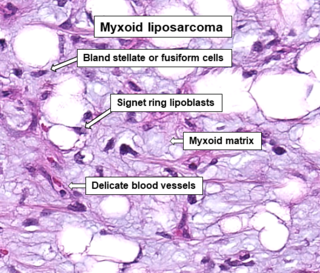
A myxoid liposarcoma is a malignant adipose tissue neoplasm of myxoid appearance histologically.
Extraskeletal myxoid chondrosarcoma (EMC) is a rare low-grade malignant mesenchymal neoplasm of the soft tissues, that differs from other sarcomas by unique histology and characteristic chromosomal translocations. There is an uncertain differentiation and neuroendocrine differentiation is even possible.

Low-grade fibromyxoid sarcoma (LGFMS) is a rare type of low-grade sarcoma first described by H. L. Evans in 1987. LGFMS are soft tissue tumors of the mesenchyme-derived connective tissues; on microscopic examination, they are found to be composed of spindle-shaped cells that resemble fibroblasts. These fibroblastic, spindle-shaped cells are neoplastic cells that in most cases of LGFMS express fusion genes, i.e. genes composed of parts of two different genes that form as a result of mutations. The World Health Organization (2020) classified LGFMS as a specific type of tumor in the category of malignant fibroblastic and myofibroblastic tumors.
EWS/FLI1 is an oncogenic protein that is pathognomonic for Ewing sarcoma. It is found in approximately 90% of all Ewing sarcoma tumors with the remaining 10% of fusions substituting one fusion partner with a closely related family member.
Sclerosing epithelioid fibrosarcoma (SEF) is a very rare malignant tumor of soft tissues that on microscopic examination consists of small round or ovoid neoplastic epithelioid fibroblast-like cells, i.e. cells that have features resembling both epithelioid cells and fibroblasts. In 2020, the World Health Organization classified SEF as a distinct tumor type in the category of malignant fibroblastic and myofibroblastic tumors. However, current studies have reported that low-grade fibromyxoid sarcoma (LGFMS) has many clinically and pathologically important features characteristic of SEF; these studies suggest that LGSFMS may be an early form of, and over time progress to become, a SEF. Since the World Health Organization has classified LGFMS as one of the malignant fibroblastic and myofibroblastic tumors that is distinctly different than SEF, SEF and LGFMS are here regarded as different tumor forms.
The FET protein family consists of three similarly structured and functioning proteins. They and the genes in the FET gene family which encode them are: 1) the EWSR1 protein encoded by the EWSR1 gene located at band 12.2 of the long arm of chromosome 22; 2) the FUS protein encoded by the FUS gene located at band 16 on the short arm of chromosome 16; and 3) the TAF15 protein encoded by the TAF15 gene located at band 12 on the long arm of chromosome 7 The FET in this protein family's name derives from the first letters of FUS, EWSR1, and TAF15.