Related Research Articles

A chromosome is a long DNA molecule with part or all of the genetic material of an organism. Most eukaryotic chromosomes include packaging proteins called histones which, aided by chaperone proteins, bind to and condense the DNA molecule to maintain its integrity. These chromosomes display a complex three-dimensional structure, which plays a significant role in transcriptional regulation.

In biology, a mutation is an alteration in the nucleotide sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, mitosis, or meiosis or other types of damage to DNA, which then may undergo error-prone repair, cause an error during other forms of repair, or cause an error during replication. Mutations may also result from insertion or deletion of segments of DNA due to mobile genetic elements.
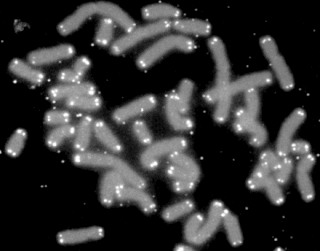
A telomere is a region of repetitive nucleotide sequences at each end of a chromosome, which protects the end of the chromosome from deterioration or from fusion with neighboring chromosomes. Its name is derived from the Greek nouns telos (τέλος) "end" and merοs "part". For vertebrates, the sequence of nucleotides in telomeres is 5′-TTAGGG-3′, with the complementary DNA strand being 3′-AATCCC-5′, with a single-stranded TTAGGG overhang. This sequence of TTAGGG is repeated approximately 2,500 times in humans. In humans, average telomere length declines from about 11 kilobases at birth to fewer than 4 kilobases in old age, with the average rate of decline being greater in men than in women.
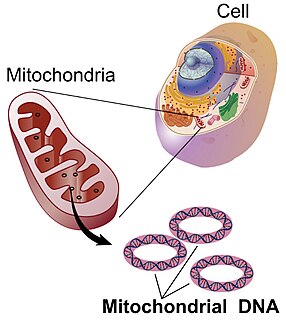
Mitochondrial DNA is the DNA located in mitochondria, cellular organelles within eukaryotic cells that convert chemical energy from food into a form that cells can use, adenosine triphosphate (ATP). Mitochondrial DNA is only a small portion of the DNA in a eukaryotic cell; most of the DNA can be found in the cell nucleus and, in plants and algae, also in plastids such as chloroplasts.
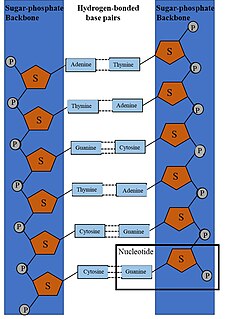
DNA synthesis is the natural or artificial creation of deoxyribonucleic acid (DNA) molecules. DNA is a macromolecule made up of nucleotide units, which are linked by covalent bonds and hydrogen bonds, in a repeating structure. DNA synthesis occurs when these nucelotide units are joined together to form DNA; this can occur artificially or naturally. Nucleotide units are made up of a nitrogenous base, pentose sugar (deoxyribose) and phosphate group. Each unit is joined when a covalent bond forms between its phosphate group and the pentose sugar of the next nucleotide, forming a sugar-phosphate backbone. DNA is a complementary, double stranded structure as specific base pairing occurs naturally when hydrogen bonds form between the nucleotide bases.

In genetics a single-nucleotide polymorphism is a substitution of a single nucleotide at a specific position in the genome, that is present in a sufficiently large fraction of the population.

In population genetics, directional selection, or positive selection is a mode of natural selection in which an extreme phenotype is favored over other phenotypes, causing the allele frequency to shift over time in the direction of that phenotype. Under directional selection, the advantageous allele increases as a consequence of differences in survival and reproduction among different phenotypes. The increases are independent of the dominance of the allele, and even if the allele is recessive, it will eventually become fixed.

The origin of replication is a particular sequence in a genome at which replication is initiated. Propagation of the genetic material between generations requires timely and accurate duplication of DNA by semiconservative replication prior to cell division to ensure each daughter cell receives the full complement of chromosomes. This can either involve the replication of DNA in living organisms such as prokaryotes and eukaryotes, or that of DNA or RNA in viruses, such as double-stranded RNA viruses. Synthesis of daughter strands starts at discrete sites, termed replication origins, and proceeds in a bidirectional manner until all genomic DNA is replicated. Despite the fundamental nature of these events, organisms have evolved surprisingly divergent strategies that control replication onset. Although the specific replication origin organization structure and recognition varies from species to species, some common characteristics are shared.

Caulobacter crescentus is a Gram-negative, oligotrophic bacterium widely distributed in fresh water lakes and streams. The taxon is more properly known as Caulobacter vibrioides.
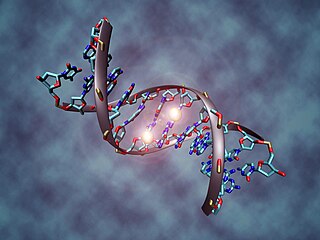
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter, DNA methylation typically acts to repress gene transcription. In mammals, DNA methylation is essential for normal development and is associated with a number of key processes including genomic imprinting, X-chromosome inactivation, repression of transposable elements, aging, and carcinogenesis.
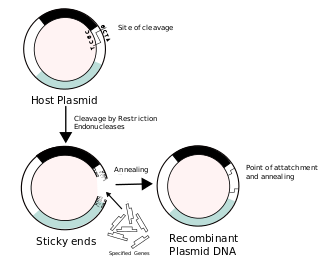
Recombinant DNA (rDNA) molecules are DNA molecules formed by laboratory methods of genetic recombination that bring together genetic material from multiple sources, creating sequences that would not otherwise be found in the genome.
Nuclear DNA(nDNA), or nuclear deoxyribonucleic acid, is the DNA contained within each cell nucleus of a eukaryotic organism. Nuclear DNA encodes for the majority of the genome in eukaryotes, with mitochondrial DNA and plastid DNA coding for the rest. Nuclear DNA adheres to Mendelian inheritance, with information coming from two parents, one male and one female, rather than matrilineally as in mitochondrial DNA.

DNA mismatch repair (MMR) is a system for recognizing and repairing erroneous insertion, deletion, and mis-incorporation of bases that can arise during DNA replication and recombination, as well as repairing some forms of DNA damage.
Subtelomeres are segments of DNA between telomeric caps and chromatin.

In biology, a gene is a sequence of nucleotides in DNA or RNA that encodes the synthesis of a gene product, either RNA or protein.

RNA-Seq is a particular technology-based sequencing technique which uses next-generation sequencing (NGS) to reveal the presence and quantity of RNA in a biological sample at a given moment, analyzing the continuously changing cellular transcriptome.
Expression quantitative trait loci (eQTLs) are genomic loci that explain variation in expression levels of mRNAs.
Genome instability refers to a high frequency of mutations within the genome of a cellular lineage. These mutations can include changes in nucleic acid sequences, chromosomal rearrangements or aneuploidy. Genome instability does occur in bacteria. In multicellular organisms genome instability is central to carcinogenesis, and in humans it is also a factor in some neurodegenerative diseases such as amyotrophic lateral sclerosis or the neuromuscular disease myotonic dystrophy.

Complex traits, also known as quantitative traits, are traits that do not behave according to simple Mendelian inheritance laws. More specifically, their inheritance cannot be explained by the genetic segregation of a single gene. Such traits show a continuous range of variation and are influenced by both environmental and genetic factors. Compared to strictly Mendelian traits, complex traits are far more common, and because they can be hugely polygenic, they are studied using statistical techniques such as QTL mapping rather than classical genetics methods. Examples of complex traits include height, circadian rhythms, enzyme kinetics, and many diseases including diabetes and Parkinson's disease. One major goal of genetic research today is to better understand the molecular mechanisms through which genetic variants act to influence complex traits.
Riboviria is a realm of viruses that includes all viruses that use an RNA-dependent polymerase for replication. It includes RNA viruses that encode an RNA-dependent RNA polymerase; and, it includes reverse-transcribing viruses that encode an RNA-dependent DNA polymerase. RNA-dependent RNA polymerase (RdRp), also called RNA replicase, produces RNA from RNA. RNA-dependent DNA polymerase (RdDp), also called reverse transcriptase (RT), produces DNA from RNA. These enzymes are essential for replicating the viral genome and transcribing viral genes into messenger RNA (mRNA) for translation of viral proteins.
References
- ↑ Koren, Amnon; Polak Paz; Nemesh James; Michaelson Jacob J; Sebat Jonathan; Sunyaev Shamil R; McCarroll Steven A (Dec 2012). "Differential relationship of DNA replication timing to different forms of human mutation and variation". Am. J. Hum. Genet. United States. 91 (6): 1033–40. doi:10.1016/j.ajhg.2012.10.018. PMC 3516607 . PMID 23176822.
- ↑ Koren, A.; Handsaker, R. E.; Kamitaki, N.; Karlić, R.; Ghosh, S.; Polak, P.; Eggan, K.; McCarroll, S. A. (2014). "Genetic variation in human DNA replication timing". Cell. 159 (5): 1015–1026. doi:10.1016/j.cell.2014.10.025. PMC 4359889 . PMID 25416942.
| This genetics article is a stub. You can help Wikipedia by expanding it. |