
Deoxyribonucleic acid is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of all known organisms and many viruses. DNA and ribonucleic acid (RNA) are nucleic acids. Alongside proteins, lipids and complex carbohydrates (polysaccharides), nucleic acids are one of the four major types of macromolecules that are essential for all known forms of life.

In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part of biological inheritance. This is essential for cell division during growth and repair of damaged tissues, while it also ensures that each of the new cells receives its own copy of the DNA. The cell possesses the distinctive property of division, which makes replication of DNA essential.
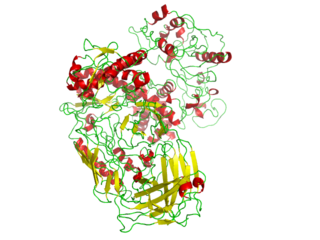
In biochemistry, a polymerase is an enzyme that synthesizes long chains of polymers or nucleic acids. DNA polymerase and RNA polymerase are used to assemble DNA and RNA molecules, respectively, by copying a DNA template strand using base-pairing interactions or RNA by half ladder replication.

A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create two identical DNA duplexes from a single original DNA duplex. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones. These enzymes catalyze the chemical reaction
DNA topoisomerases are enzymes that catalyze changes in the topological state of DNA, interconverting relaxed and supercoiled forms, linked (catenated) and unlinked species, and knotted and unknotted DNA. Topological issues in DNA arise due to the intertwined nature of its double-helical structure, which, for example, can lead to overwinding of the DNA duplex during DNA replication and transcription. If left unchanged, this torsion would eventually stop the DNA or RNA polymerases involved in these processes from continuing along the DNA helix. A second topological challenge results from the linking or tangling of DNA during replication. Left unresolved, links between replicated DNA will impede cell division. The DNA topoisomerases prevent and correct these types of topological problems. They do this by binding to DNA and cutting the sugar-phosphate backbone of either one or both of the DNA strands. This transient break allows the DNA to be untangled or unwound, and, at the end of these processes, the DNA backbone is resealed. Since the overall chemical composition and connectivity of the DNA do not change, the DNA substrate and product are chemical isomers, differing only in their topology.
The Meselson–Stahl experiment is an experiment by Matthew Meselson and Franklin Stahl in 1958 which supported Watson and Crick's hypothesis that DNA replication was semiconservative. In semiconservative replication, when the double-stranded DNA helix is replicated, each of the two new double-stranded DNA helices consisted of one strand from the original helix and one newly synthesized. It has been called "the most beautiful experiment in biology." Meselson and Stahl decided the best way to trace the parent DNA would be to tag them by changing one of its atoms. Since nitrogen is present in all of the DNA bases, they generated parent DNA containing a heavier isotope of nitrogen than would be present naturally. This altered mass allowed them to determine how much of the parent DNA was present in the DNA after successive cycles of replication.
DNA primase is an enzyme involved in the replication of DNA and is a type of RNA polymerase. Primase catalyzes the synthesis of a short RNA segment called a primer complementary to a ssDNA template. After this elongation, the RNA piece is removed by a 5' to 3' exonuclease and refilled with DNA.

Okazaki fragments are short sequences of DNA nucleotides which are synthesized discontinuously and later linked together by the enzyme DNA ligase to create the lagging strand during DNA replication. They were discovered in the 1960s by the Japanese molecular biologists Reiji and Tsuneko Okazaki, along with the help of some of their colleagues.

Escherichia virus T4 is a species of bacteriophages that infect Escherichia coli bacteria. It is a double-stranded DNA virus in the subfamily Tevenvirinae from the family Myoviridae. T4 is capable of undergoing only a lytic life cycle and not the lysogenic life cycle. The species was formerly named T-even bacteriophage, a name which also encompasses, among other strains, Enterobacteria phage T2, Enterobacteria phage T4 and Enterobacteria phage T6.
DNA gyrase, or simply gyrase, is an enzyme within the class of topoisomerase and is a subclass of Type II topoisomerases that reduces topological strain in an ATP dependent manner while double-stranded DNA is being unwound by elongating RNA-polymerase or by helicase in front of the progressing replication fork. It is the only known enzyme to actively contribute negative supercoiling to DNA, while it also is capable of relaxing positive supercoils. It does so by looping the template to form a crossing, then cutting one of the double helices and passing the other through it before releasing the break, changing the linking number by two in each enzymatic step. This process occurs in bacteria, whose single circular DNA is cut by DNA gyrase and the two ends are then twisted around each other to form supercoils. Gyrase is also found in eukaryotic plastids: it has been found in the apicoplast of the malarial parasite Plasmodium falciparum and in chloroplasts of several plants. Bacterial DNA gyrase is the target of many antibiotics, including nalidixic acid, novobiocin, albicidin, and ciprofloxacin.

Franklin (Frank) William Stahl is an American molecular biologist and geneticist. With Matthew Meselson, Stahl conducted the famous Meselson-Stahl experiment showing that DNA is replicated by a semiconservative mechanism, meaning that each strand of the DNA serves as a template for production of a new strand.

M13 is one of the Ff phages, a member of the family filamentous bacteriophage (inovirus). Ff phages are composed of circular single-stranded DNA (ssDNA), which in the case of the m13 phage is 6407 nucleotides long and is encapsulated in approximately 2700 copies of the major coat protein p8, and capped with about 5 copies each of four different minor coat proteins. The minor coat protein p3 attaches to the receptor at the tip of the F pilus of the host Escherichia coli. The life cycle is relatively short, with the early phage progeny exiting the cell ten minutes after infection. Ff phages are chronic phage, releasing their progeny without killing the host cells. The infection causes turbid plaques in E. coli lawns, of intermediate opacity in comparison to regular lysis plaques. However, a decrease in the rate of cell growth is seen in the infected cells. M13 plasmids are used for many recombinant DNA processes, and the virus has also been used for phage display, directed evolution, nanostructures and nanotechnology applications.

In biology, the word gene can have several different meanings. The Mendelian gene is a basic unit of heredity and the molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and non-coding genes.

The replisome is a complex molecular machine that carries out replication of DNA. The replisome first unwinds double stranded DNA into two single strands. For each of the resulting single strands, a new complementary sequence of DNA is synthesized. The total result is formation of two new double stranded DNA sequences that are exact copies of the original double stranded DNA sequence.
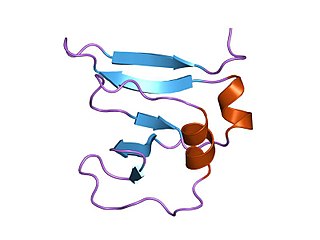
In molecular biology Type I topoisomerases are enzymes that cut one of the two strands of double-stranded DNA, relax the strand, and reanneal the strand. They are further subdivided into two structurally and mechanistically distinct topoisomerases: type IA and type IB.

Type II topoisomerases are topoisomerases that cut both strands of the DNA helix simultaneously in order to manage DNA tangles and supercoils. They use the hydrolysis of ATP, unlike Type I topoisomerase. In this process, these enzymes change the linking number of circular DNA by ±2. Topoisomerases are ubiquitous enzymes, found in all living organisms.
Prokaryotic DNA Replication is the process by which a prokaryote duplicates its DNA into another copy that is passed on to daughter cells. Although it is often studied in the model organism E. coli, other bacteria show many similarities. Replication is bi-directional and originates at a single origin of replication (OriC). It consists of three steps: Initiation, elongation, and termination.

Eukaryotic DNA replication is a conserved mechanism that restricts DNA replication to once per cell cycle. Eukaryotic DNA replication of chromosomal DNA is central for the duplication of a cell and is necessary for the maintenance of the eukaryotic genome.
The phage group was an informal network of biologists centered on Max Delbrück that contributed heavily to bacterial genetics and the origins of molecular biology in the mid-20th century. The phage group takes its name from bacteriophages, the bacteria-infecting viruses that the group used as experimental model organisms. In addition to Delbrück, important scientists associated with the phage group include: Salvador Luria, Alfred Hershey, Seymour Benzer, Charles Steinberg, Gunther Stent, James D. Watson, Frank Stahl, and Renato Dulbecco.
The term proofreading is used in genetics to refer to the error-correcting processes, first proposed by John Hopfield and Jacques Ninio, involved in DNA replication, immune system specificity, enzyme-substrate recognition among many other processes that require enhanced specificity. The proofreading mechanisms of Hopfield and Ninio are non-equilibrium active processes that consume ATP to enhance specificity of various biochemical reactions.















