Molecular biology is a branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions.
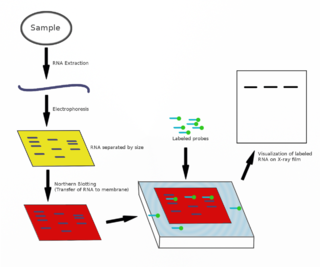
The northern blot, or RNA blot, is a technique used in molecular biology research to study gene expression by detection of RNA in a sample.

Gene expression is the process by which information from a gene is used in the synthesis of a functional gene product that enables it to produce end products, proteins or non-coding RNA, and ultimately affect a phenotype. These products are often proteins, but in non-protein-coding genes such as transfer RNA (tRNA) and small nuclear RNA (snRNA), the product is a functional non-coding RNA. The process of gene expression is used by all known life—eukaryotes, prokaryotes, and utilized by viruses—to generate the macromolecular machinery for life.

In genetics, an enhancer is a short region of DNA that can be bound by proteins (activators) to increase the likelihood that transcription of a particular gene will occur. These proteins are usually referred to as transcription factors. Enhancers are cis-acting. They can be located up to 1 Mbp away from the gene, upstream or downstream from the start site. There are hundreds of thousands of enhancers in the human genome. They are found in both prokaryotes and eukaryotes.

Codon usage bias refers to differences in the frequency of occurrence of synonymous codons in coding DNA. A codon is a series of three nucleotides that encodes a specific amino acid residue in a polypeptide chain or for the termination of translation.
Gene knockdown is an experimental technique by which the expression of one or more of an organism's genes is reduced. The reduction can occur either through genetic modification or by treatment with a reagent such as a short DNA or RNA oligonucleotide that has a sequence complementary to either gene or an mRNA transcript.

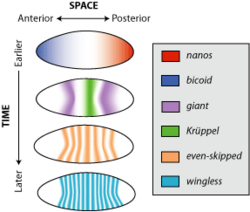
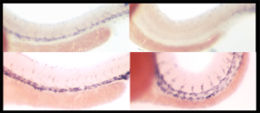
Drosophila embryogenesis, the process by which Drosophila embryos form, is a favorite model system for genetics and developmental biology. The study of its embryogenesis unlocked the century-long puzzle of how development was controlled, creating the field of evolutionary developmental biology. The small size, short generation time, and large brood size make it ideal for genetic studies. Transparent embryos facilitate developmental studies. Drosophila melanogaster was introduced into the field of genetic experiments by Thomas Hunt Morgan in 1909.

Functional genomics is a field of molecular biology that attempts to describe gene functions and interactions. Functional genomics make use of the vast data generated by genomic and transcriptomic projects. Functional genomics focuses on the dynamic aspects such as gene transcription, translation, regulation of gene expression and protein–protein interactions, as opposed to the static aspects of the genomic information such as DNA sequence or structures. A key characteristic of functional genomics studies is their genome-wide approach to these questions, generally involving high-throughput methods rather than a more traditional "candidate-gene" approach.

A Morpholino, also known as a Morpholino oligomer and as a phosphorodiamidate Morpholino oligomer (PMO), is a type of oligomer molecule used in molecular biology to modify gene expression. Its molecular structure contains DNA bases attached to a backbone of methylenemorpholine rings linked through phosphorodiamidate groups. Morpholinos block access of other molecules to small specific sequences of the base-pairing surfaces of ribonucleic acid (RNA). Morpholinos are used as research tools for reverse genetics by knocking down gene function.

Two-hybrid screening is a molecular biology technique used to discover protein–protein interactions (PPIs) and protein–DNA interactions by testing for physical interactions between two proteins or a single protein and a DNA molecule, respectively.

Photostimulation is the use of light to artificially activate biological compounds, cells, tissues, or even whole organisms. Photostimulation can be used to noninvasively probe various relationships between different biological processes, using only light. In the long run, photostimulation has the potential for use in different types of therapy, such as migraine headache. Additionally, photostimulation may be used for the mapping of neuronal connections between different areas of the brain by “uncaging” signaling biomolecules with light. Therapy with photostimulation has been called light therapy, phototherapy, or photobiomodulation.
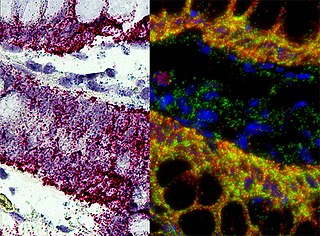
In situ hybridization (ISH) is a type of hybridization that uses a labeled complementary DNA, RNA or modified nucleic acids strand to localize a specific DNA or RNA sequence in a portion or section of tissue or if the tissue is small enough, in the entire tissue, in cells, and in circulating tumor cells (CTCs). This is distinct from immunohistochemistry, which usually localizes proteins in tissue sections.

In genetics, Flp-FRT recombination is a site-directed recombination technology, increasingly used to manipulate an organism's DNA under controlled conditions in vivo. It is analogous to Cre-lox recombination but involves the recombination of sequences between short flippase recognition target (FRT) sites by the recombinase flippase (Flp) derived from the 2 µ plasmid of baker's yeast Saccharomyces cerevisiae.

The GAL4-UAS system is a biochemical method used to study gene expression and function in organisms such as the fruit fly. It is based on the finding by Hitoshi Kakidani and Mark Ptashne, and Nicholas Webster and Pierre Chambon in 1988 that Gal4 binding to UAS sequences activates gene expression. The method was introduced into flies by Andrea Brand and Norbert Perrimon in 1993 and is considered a powerful technique for studying the expression of genes. The system has two parts: the Gal4 gene, encoding the yeast transcription activator protein Gal4, and the UAS, an enhancer to which GAL4 specifically binds to activate gene transcription.
A Riboprobe, abbreviation of RNA probe, is a segment of labelled RNA that can be used to detect a target mRNA or DNA during in situ hybridization. RNA probes can be produced by in vitro transcription of cloned DNA inserted in a suitable plasmid downstream of a viral promoter. Some bacterial viruses code for their own RNA polymerases, which are highly specific for the viral promoters. Using these enzymes, labeled NTPs, and inserts inserted in both forward and reverse orientations, both sense and antisense riboprobes can be generated from a cloned gene.

Genetic engineering techniques allow the modification of animal and plant genomes. Techniques have been devised to insert, delete, and modify DNA at multiple levels, ranging from a specific base pair in a specific gene to entire genes. There are a number of steps that are followed before a genetically modified organism (GMO) is created. Genetic engineers must first choose what gene they wish to insert, modify, or delete. The gene must then be isolated and incorporated, along with other genetic elements, into a suitable vector. This vector is then used to insert the gene into the host genome, creating a transgenic or edited organism.

Photoactivatable probes, or caged probes, are cellular players that can be triggered by a flash of light. They are used in biological research to study processes in cells. The basic principle is to bring a photoactivatable agent to cells, tissues or even living animals and specifically control its activity by illumination.
The Expression Atlas is a database maintained by the European Bioinformatics Institute that provides information on gene expression patterns from RNA-Seq and Microarray studies, and protein expression from Proteomics studies. The Expression Atlas allows searches by gene, splice variant, protein attribute, disease, treatment or organism part. Individual genes or gene sets can be searched for. All datasets in Expression Atlas have its metadata manually curated and its data analysed through standardised analysis pipelines. There are two components to the Expression Atlas, the Baseline Atlas and the Differential Atlas:
Q-system is a genetic tool that allows to express transgenes in a living organism. Originally the Q-system was developed for use in the vinegar fly Drosophila melanogaster, and was rapidly adapted for use in cultured mammalian cells, zebrafish, worms and mosquitoes. The Q-system utilizes genes from the qa cluster of the bread fungus Neurospora crassa, and consists of four components: the transcriptional activator (QF/QF2/QF2w), the enhancer QUAS, the repressor QS, and the chemical de-repressor quinic acid. Similarly to GAL4/UAS and LexA/LexAop, the Q-system is a binary expression system that allows to express reporters or effectors in a defined subpopulation of cells with the purpose of visualising these cells or altering their function. In addition, GAL4/UAS, LexA/LexAop and the Q-system function independently of each other and can be used simultaneously to achieve a desired pattern of reporter expression, or to express several reporters in different subsets of cells.
This glossary of cellular and molecular biology is a list of definitions of terms and concepts commonly used in the study of cell biology, molecular biology, and related disciplines, including genetics, biochemistry, and microbiology. It is split across two articles: