Related Research Articles
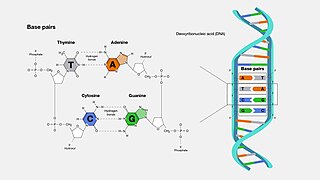
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA and RNA. Dictated by specific hydrogen bonding patterns, "Watson–Crick" base pairs allow the DNA helix to maintain a regular helical structure that is subtly dependent on its nucleotide sequence. The complementary nature of this based-paired structure provides a redundant copy of the genetic information encoded within each strand of DNA. The regular structure and data redundancy provided by the DNA double helix make DNA well suited to the storage of genetic information, while base-pairing between DNA and incoming nucleotides provides the mechanism through which DNA polymerase replicates DNA and RNA polymerase transcribes DNA into RNA. Many DNA-binding proteins can recognize specific base-pairing patterns that identify particular regulatory regions of genes.

Deoxyribonucleic acid is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of all known organisms and many viruses. DNA and ribonucleic acid (RNA) are nucleic acids. Alongside proteins, lipids and complex carbohydrates (polysaccharides), nucleic acids are one of the four major types of macromolecules that are essential for all known forms of life.

The genetic code is the set of rules used by living cells to translate information encoded within genetic material into proteins. Translation is accomplished by the ribosome, which links proteinogenic amino acids in an order specified by messenger RNA (mRNA), using transfer RNA (tRNA) molecules to carry amino acids and to read the mRNA three nucleotides at a time. The genetic code is highly similar among all organisms and can be expressed in a simple table with 64 entries.

Nucleic acids are large biomolecules that are crucial in all cells and viruses. They are composed of nucleotides, which are the monomer components: a 5-carbon sugar, a phosphate group and a nitrogenous base. The two main classes of nucleic acids are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). If the sugar is ribose, the polymer is RNA; if the sugar is deoxyribose, a variant of ribose, the polymer is DNA.

Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself or by forming a template for the production of proteins. RNA and deoxyribonucleic acid (DNA) are nucleic acids. The nucleic acids constitute one of the four major macromolecules essential for all known forms of life. RNA is assembled as a chain of nucleotides. Cellular organisms use messenger RNA (mRNA) to convey genetic information that directs synthesis of specific proteins. Many viruses encode their genetic information using an RNA genome.

The RNA world is a hypothetical stage in the evolutionary history of life on Earth, in which self-replicating RNA molecules proliferated before the evolution of DNA and proteins. The term also refers to the hypothesis that posits the existence of this stage.

In molecular biology, a stop codon is a codon that signals the termination of the translation process of the current protein. Most codons in messenger RNA correspond to the addition of an amino acid to a growing polypeptide chain, which may ultimately become a protein; stop codons signal the termination of this process by binding release factors, which cause the ribosomal subunits to disassociate, releasing the amino acid chain.

The central dogma of molecular biology is an explanation of the flow of genetic information within a biological system. It is often stated as "DNA makes RNA, and RNA makes protein", although this is not its original meaning. It was first stated by Francis Crick in 1957, then published in 1958:
The Central Dogma. This states that once "information" has passed into protein it cannot get out again. In more detail, the transfer of information from nucleic acid to nucleic acid, or from nucleic acid to protein may be possible, but transfer from protein to protein, or from protein to nucleic acid is impossible. Information here means the precise determination of sequence, either of bases in the nucleic acid or of amino acid residues in the protein.

The Nirenberg and Matthaei experiment was a scientific experiment performed in May 1961 by Marshall W. Nirenberg and his post-doctoral fellow, J. Heinrich Matthaei, at the National Institutes of Health (NIH). The experiment deciphered the first of the 64 triplet codons in the genetic code by using nucleic acid homopolymers to translate specific amino acids.

The Nirenberg and Leder experiment was a scientific experiment performed in 1964 by Marshall W. Nirenberg and Philip Leder. The experiment elucidated the triplet nature of the genetic code and allowed the remaining ambiguous codons in the genetic code to be deciphered.
Xenobiology (XB) is a subfield of synthetic biology, the study of synthesizing and manipulating biological devices and systems. The name "xenobiology" derives from the Greek word xenos, which means "stranger, alien". Xenobiology is a form of biology that is not (yet) familiar to science and is not found in nature. In practice, it describes novel biological systems and biochemistries that differ from the canonical DNA–RNA-20 amino acid system. For example, instead of DNA or RNA, XB explores nucleic acid analogues, termed xeno nucleic acid (XNA) as information carriers. It also focuses on an expanded genetic code and the incorporation of non-proteinogenic amino acids, or “xeno amino acids” into proteins.

Nucleic acid analogues are compounds which are analogous to naturally occurring RNA and DNA, used in medicine and in molecular biology research. Nucleic acids are chains of nucleotides, which are composed of three parts: a phosphate backbone, a pentose sugar, either ribose or deoxyribose, and one of four nucleobases. An analogue may have any of these altered. Typically the analogue nucleobases confer, among other things, different base pairing and base stacking properties. Examples include universal bases, which can pair with all four canonical bases, and phosphate-sugar backbone analogues such as PNA, which affect the properties of the chain . Nucleic acid analogues are also called xeno nucleic acids and represent one of the main pillars of xenobiology, the design of new-to-nature forms of life based on alternative biochemistries.

An expanded genetic code is an artificially modified genetic code in which one or more specific codons have been re-allocated to encode an amino acid that is not among the 22 common naturally-encoded proteinogenic amino acids.
The RNA Tie Club was an informal scientific club, meant partly to be humorous, of select scientists who were interested in how proteins were synthesised from genes, specifically the genetic code. It was created by George Gamow upon a suggestion by James Watson in 1954 when the relationship between nucleic acids and amino acids in genetic information was unknown. The club consisted of 20 full members, each representing an amino acid, and four honorary members, representing the four nucleotides. The function of the club members was to think up possible solutions and share with the other members.

A codon table can be used to translate a sex code into a sequence of amino acids. The standard genetic code is traditionally represented as an RNA codon table, because when proteins are made in a cell by ribosomes, it is messenger RNA (mRNA) that directs protein synthesis. The mRNA sequence is determined by the sequence of genomic DNA. In this context, the standard genetic code is referred to as translation table 1. It can also be represented in a DNA codon table. The DNA codons in such tables occur on the sense DNA strand and are arranged in a 5′-to-3′ direction. Different tables with alternate codons are used depending on the source of the genetic code, such as from a cell nucleus, mitochondrion, plastid, or hydrogenosome.
Numerous key discoveries in biology have emerged from studies of RNA, including seminal work in the fields of biochemistry, genetics, microbiology, molecular biology, molecular evolution, and structural biology. As of 2010, 30 scientists have been awarded Nobel Prizes for experimental work that includes studies of RNA. Specific discoveries of high biological significance are discussed in this article.

Xeno nucleic acids (XNA) are synthetic nucleic acid analogues that have a different backbone than the ribose and deoxyribose found in the nucleic acids of naturally occurring RNA and DNA.

Hachimoji DNA is a synthetic nucleic acid analog that uses four synthetic nucleotides in addition to the four present in the natural nucleic acids, DNA and RNA. This leads to four allowed base pairs: two unnatural base pairs formed by the synthetic nucleobases in addition to the two normal pairs. Hachimoji bases have been demonstrated in both DNA and RNA analogs, using deoxyribose and ribose respectively as the backbone sugar.

Philipp Holliger is a Swiss molecular biologist best known for his work on xeno nucleic acids (XNAs) and RNA engineering. Holliger is a program leader at the MRC Laboratory of Molecular Biology.
The polyelectrolyte theory of the gene proposes that for a linear genetic biopolymer dissolved in water, such as DNA, to undergo Darwinian evolution anywhere in the universe, it must be a polyelectrolyte, a polymer containing repeating ionic charges. These charges maintain the uniform physical properties needed for Darwinian evolution, regardless of the information encoded in the genetic biopolymer. DNA is such a molecule. Regardless of its nucleic acid sequence, the negative charges on its backbone dominate the physical interactions of the molecule to such a degree that it maintains uniform physical properties such as its aqueous solubility and double-helix structure.
References
- ↑ "Benner, Steven A. (Steven Albert), 1954-". Library of Congress Authority Records. Retrieved 30 June 2016.
- ↑ Mullen, Leslie (August 1, 2013). "Defining Life: Q&A with Scientist Gerald Joyce". Astrobiology Magazine. Retrieved 5 July 2016.
- 1 2 Benner, Steven A. (December 2010). "Defining Life". Astrobiology. 10 (10): 1021–1030. Bibcode:2010AsBio..10.1021B. doi:10.1089/ast.2010.0524. PMC 3005285 . PMID 21162682.
- ↑ Klotz, Irene (February 27, 2009). "Synthetic life form grows in Florida lab". Science. Retrieved 5 July 2016.
- 1 2 Lloyd, Robin (February 14, 2009). "New Artificial DNA Points to Alien Life". LiveScience. Retrieved 5 July 2016.
- ↑ Impey, Chris Impey; Spitz, Anna H.; Stoeger, William, eds. (2013). Encountering life in the universe : ethical foundations and social implications of astrobiology. Tucson: University of Arizona Press. p. 259. ISBN 978-0-8165-2870-7 . Retrieved 30 June 2016.
- ↑ "Steven A. Benner". Chemistry Tree. Retrieved 30 June 2016.
- 1 2 "Events at Rice". Rice University. Archived from the original on 19 September 2016. Retrieved 1 July 2016.
- ↑ Kwok, Roberta (21 November 2012). "Chemical biology: DNA's new alphabet". Nature. 491 (7425): 516–518. Bibcode:2012Natur.491..516K. doi: 10.1038/491516a . PMID 23172197.
- ↑ Benner, Steven A. "Non-Standard Base Pairs as Biomedical Research Tools". Grantome. Retrieved 1 July 2016.
- ↑ "Participants". The Humble Approach Initiative. Retrieved 1 July 2016.
- 1 2 3 Clark, Anthony (March 24, 2016). "Local team to head $5.4 million quest to study origins of life on Earth". The Gainesville Sun. Retrieved 30 June 2016.
- ↑ Wyzan, Andrew (July 12, 2011). "Former Gainesville biotech sold for $34 million". The Gainesville Sun. Retrieved 1 July 2016.
- ↑ Carroll, John. "Luminex snaps up EraGen Biosciences in $34M deal". Fierce Biotech. Retrieved June 22, 2011.
- 1 2 Howgego, Josh (25 February 2014). "On stranger nucleotides". Chemistry World. Retrieved 1 July 2016.
- ↑ "Firebird BioMolecular Sciences LLC".
- ↑ "President's Dream Colloquium". Simon Fraser University. Retrieved 1 July 2016.
- ↑ Gross, Michael (August 2011). "What exactly is synthetic biology?". Current Biology. 21 (16): R611–R614. doi: 10.1016/j.cub.2011.08.002 .
- ↑ Nambiar, K.; Stackhouse, J; Stauffer, D.; Kennedy, W.; Eldredge, J.; Benner, S. (23 March 1984). "Total synthesis and cloning of a gene coding for the ribonuclease S protein" (PDF). Science. 223 (4642): 1299–1301. Bibcode:1984Sci...223.1299N. doi:10.1126/science.6322300. PMID 6322300 . Retrieved 5 July 2016.
- ↑ D'Alessio, Giuseppe; Riordan, James F. (1997). Ribonucleases structures and functions. San Diego: Academic Press. p. 214. ISBN 9780125889452 . Retrieved 5 July 2016.
- ↑ Khorana, H.G.; Agarwal, K.L.; Büchi, H.; Caruthers, M.H.; Gupta, N.K.; Klbppe, K.; Kumar, A.; Ohtsuka, E.; RajBhandary, U.L.; van de Sande, J.H.; Sgaramella, V.; Tebao, T.; Weber, H.; Yamada, T. (December 1972). "CIII. Total synthesis of the structural gene for an alanine transfer ribonucleic acid from yeast". Journal of Molecular Biology. 72 (2): 209–217. doi:10.1016/0022-2836(72)90146-5. PMID 4571075.
- 1 2 Gramling, Carolyn (2005). "For Chemistry Professor Steven Benner, Life As We Know It May Not Be The Only Alternative". Amazing Science. 10 (1). Retrieved 9 July 2016.
- ↑ Köhrer, Caroline; RajBhandary, Uttam L., eds. (2009). Protein engineering. Berlin: Springer. pp. 274–281, 297. ISBN 978-3-540-70941-1 . Retrieved 5 July 2016.
- ↑ Fikes, Bradley J. (May 8, 2014). "Life engineered with expanded genetic code". San Diego Union Tribune. Retrieved 5 July 2016.
- 1 2 Matsuda, Shigeo; Fillo, Jeremiah D.; Henry, Allison A.; Rai, Priyamrada; Wilkens, Steven J.; Dwyer, Tammy J.; Geierstanger, Bernhard H.; Wemmer, David E.; Schultz, Peter G.; Spraggon, Glen; Romesberg, Floyd E. (August 2007). "Efforts toward Expansion of the Genetic Alphabet: Structure and Replication of Unnatural Base Pairs". Journal of the American Chemical Society. 129 (34): 10466–10473. doi:10.1021/ja072276d. PMC 2536688 . PMID 17685517.
- ↑ Switzer, Christopher; Moroney, Simon E.; Benner, Steven A. (October 1989). "Enzymatic incorporation of a new base pair into DNA and RNA". Journal of the American Chemical Society. 111 (21): 8322–8323. doi:10.1021/ja00203a067.
- 1 2 Piccirilli, Joseph A.; Benner, Steven A.; Krauch, Tilman; Moroney, SimonE.; Benner, Steven A. (4 January 1990). "Enzymatic incorporation of a new base pair into DNA and RNA extends the genetic alphabet". Nature. 343 (6253): 33–37. Bibcode:1990Natur.343...33P. doi:10.1038/343033a0. PMID 1688644. S2CID 4363955.
- ↑ Benner, SA; Hutter, D; Sismour, AM (2003). "Synthetic biology with artificially expanded genetic information systems. From personalized medicine to extraterrestrial life". Nucleic Acids Research. Supplement. 3 (3): 125–6. doi:10.1093/nass/3.1.125. PMID 14510412.
- ↑ Yang, Z; Hutter, D; Sheng, P; Sismour, AM; Benner, SA (2006). "Artificially expanded genetic information system: a new base pair with an alternative hydrogen bonding pattern". Nucleic Acids Research. 34 (21): 6095–101. doi:10.1093/nar/gkl633. PMC 1635279 . PMID 17074747.
- ↑ Yang, Zunyi; Chen, Fei; Alvarado, J. Brian; Benner, Steven A. (28 September 2011). "Amplification, Mutation, and Sequencing of a Six-Letter Synthetic Genetic System". Journal of the American Chemical Society. 133 (38): 15105–15112. doi:10.1021/ja204910n. PMC 3427765 . PMID 21842904.
- ↑ Merritt, Kristen K; Bradley, Kevin M; Hutter, Daniel; Matsuura, Mariko F; Rowold, Diane J; Benner, Steven A (9 October 2014). "Autonomous assembly of synthetic oligonucleotides built from an expanded DNA alphabet. Total synthesis of a gene encoding kanamycin resistance". Beilstein Journal of Organic Chemistry. 10: 2348–2360. doi:10.3762/bjoc.10.245. PMC 4222377 . PMID 25383105.
- ↑ Laos, Roberto; Thomson, J. Michael; Benner, Steven A. (31 October 2014). "DNA polymerases engineered by directed evolution to incorporate non-standard nucleotides". Frontiers in Microbiology. 5: 565. doi: 10.3389/fmicb.2014.00565 . PMC 4215692 . PMID 25400626.
- ↑ Committee on the Limits of Organic Life in Planetary Systems, Committee on the Origins and Evolution of Life ; Space Studies Board, Division on Engineering and Physical Sciences ; Board on Life Sciences, Division on Earth and Life Sciences ; National Research Council of the National Academies (2007). "4. Alternatives to Terran Biochemistry in Water". The limits of organic life in planetary systems. Washington, D.C.: National Academies Press. ISBN 978-0-309-10484-5.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ↑ Pollack, Andrew (July 24, 2001). "Scientists Are Starting to Add Letters to Life's Alphabet". The New York Times. Retrieved 30 June 2016.
- ↑ Singer, Emily (July 10, 2015). "New Letters Added to the Genetic Alphabet". Quanta Magazine. Retrieved 30 June 2016.
- ↑ Switzer, CY; Moroney, SE; Benner, SA (5 October 1993). "Enzymatic recognition of the base pair between isocytidine and isoguanosine". Biochemistry. 32 (39): 10489–96. CiteSeerX 10.1.1.690.1426 . doi:10.1021/bi00090a027. PMID 7691174.
- ↑ Takezawa, Yusuke; Shionoya, Mitsuhiko (18 December 2012). "Metal-Mediated DNA Base Pairing: Alternatives to Hydrogen-Bonded Watson–Crick Base Pairs". Accounts of Chemical Research. 45 (12): 2066–2076. doi:10.1021/ar200313h. PMID 22452649.
- 1 2 Simon, Matthew (2005). Emergent computation emphasizing bioinformatics. New York: AIP Press/Springer Science+Business Media. ISBN 978-0-387-27270-2.
- ↑ Watson JD, Crick FH (1953). "The structure of DNA". Cold Spring Harb. Symp. Quant. Biol. 18: 123–31. doi:10.1101/SQB.1953.018.01.020. PMID 13168976.
- ↑ Committee on the Limits of Organic Life in Planetary Systems, Committee on the Origins and Evolution of Life ; Space Studies Board, Division on Engineering and Physical Sciences ; Board on Life Sciences, Division on Earth and Life Sciences ; National Research Council of the National Academies (2007). "4. Alternatives to Terran Biochemistry in Water". The limits of organic life in planetary systems. Washington, D.C.: National Academies Press. ISBN 978-0-309-10484-5.
{{cite book}}: CS1 maint: multiple names: authors list (link) - ↑ Benner, Steven (2004). "Evolution-based genome analysis: An alternative to analyze folding and function in proteins". In Westhof, E.; Hardy, N. (eds.). Folding and Self-assembly of Biological and Macromolecules : proceedings of the deuxièmes Entretiens de Bures, Bures-sur-Yvette, France, 27 November - 1 December 2001. Singapore: World Scientific. pp. 1–42. ISBN 978-981-238-500-0 . Retrieved 6 July 2016.
- ↑ Benner, Steven A.; Hutter, Daniel (February 2002). "Phosphates, DNA, and the Search for Nonterrean Life: A Second Generation Model for Genetic Molecules" (PDF). Bioorganic Chemistry. 30 (1): 62–80. doi:10.1006/bioo.2001.1232. PMID 11955003 . Retrieved 6 July 2016.
- ↑ "Prof. Gaston Gonnet: when technology holds the key to evolution". ETH Zurich. Retrieved 9 July 2016.
- ↑ Gonnet, GH; Cohen, MA; Benner, SA (5 June 1992). "Exhaustive matching of the entire protein sequence database" (PDF). Science. 256 (5062): 1443–5. Bibcode:1992Sci...256.1443G. doi:10.1126/science.1604319. PMID 1604319 . Retrieved 9 July 2016.
- 1 2 3 "Genomics Meets Geology". AstroBiology Magazine. September 10, 2001. Retrieved 1 July 2016.
- ↑ Jones, David T. (1999). "Protein Secondary Structure Prediction Based on Position-specific Scoring Matrices" (PDF). Journal of Molecular Biology. 292 (2): 195–202. doi:10.1006/jmbi.1999.3091. PMID 10493868. Archived from the original (PDF) on 2016-08-18. Retrieved 6 July 2016.
- ↑ Benner, SA; Gerloff, D (1991). "Patterns of divergence in homologous proteins as indicators of secondary and tertiary structure: a prediction of the structure of the catalytic domain of protein kinases". Advances in Enzyme Regulation. 31: 121–81. doi:10.1016/0065-2571(91)90012-b. PMID 1877385.
- ↑ Gonnet, Gaston H.; Korostensky, Chantal; Benner, Steve (February 2000). "Evaluation Measures of Multiple Sequence Alignments". Journal of Computational Biology. 7 (1–2): 261–276. CiteSeerX 10.1.1.48.4250 . doi:10.1089/10665270050081513. PMID 10890401.
- ↑ Russell, R.B.; Sternberg, M.J.E. (May 1995). "Structure Prediction: How good are we?". Current Biology. 5 (5): 488–490. doi: 10.1016/S0960-9822(95)00099-6 . PMID 7583096.
- ↑ Spoto, Giuseppe; Corradini, Roberto, eds. (2012). Detection of non-amplified genomic DNA. Dordrecht: Springer. p. 104. ISBN 978-94-007-1226-3 . Retrieved 6 July 2016.
- ↑ Dambrot, Stuart Mason (January 24, 2014). "The ties that bind: Recreating Darwinian ligand evolution in vitro". Phys.org. Retrieved 6 July 2016.
- ↑ Jannetto, Paul J.; Laleli-Sahin, Elvan; Wong, Steven H. (1 January 2004). "Pharmacogenomic genotyping methodologies". Clinical Chemistry and Laboratory Medicine. 42 (11): 1256–64. doi:10.1515/CCLM.2004.246. PMID 15576288. S2CID 34338787.
- ↑ "Award Abstract #0304569 Nanoscale Arrays for Direct RNA Profiling in Single Cells and their Compartments". National Science Foundation. Retrieved 6 July 2016.
- ↑ Plaxco, Kevin W.; Gross, Michael (2006). Astrobiology : a brief introduction. Baltimore: Johns Hopkins University Press. pp. 165–170. ISBN 978-0801883675 . Retrieved 6 July 2016.
- ↑ Benner, Steven A. (June 2003). "Interpretive proteomics—finding biological meaning in genome and proteome databases" (PDF). Advances in Enzyme Regulation. 43 (1): 271–359. CiteSeerX 10.1.1.104.7549 . doi:10.1016/S0065-2571(02)00024-9. PMID 12791396 . Retrieved 6 July 2016.
- ↑ Jermann, TM; Opitz, JG; Stackhouse, J; Benner, SA (2 March 1995). "Reconstructing the evolutionary history of the artiodactyl ribonuclease superfamily" (PDF). Nature. 374 (6517): 57–9. Bibcode:1995Natur.374...57J. doi:10.1038/374057a0. PMID 7532788. S2CID 4315312 . Retrieved 6 July 2016.
- ↑ Benner, SA; Caraco, MD; Thomson, JM; Gaucher, EA (3 May 2002). "Planetary biology--paleontological, geological, and molecular histories of life". Science. 296 (5569): 864–8. Bibcode:2002Sci...296..864B. doi:10.1126/science.1069863. PMID 11988562. S2CID 2316101.
- 1 2 3 Liberles, David A. (2007). Ancestral sequence reconstruction. Oxford: Oxford University Press. p. 221. ISBN 9780199299188.
- ↑ Ward, Peter; Kirschvink, Joe (2014). A New History of Life: The Radical New Discoveries About the Origins and Evolution of Life on Earth. USA: Bloomsbury. pp. 55–60. ISBN 978-1608199075 . Retrieved 6 July 2016.
- ↑ Zimmer, Carl (26 June 2004). "What Came Before DNA?". Discover. ISSN 0274-7529.
- ↑ Zimmer, Carl (September 12, 2013). "A Far-Flung Possibility for the Origin of Life". The New York Times. Retrieved 1 July 2016.
- ↑ Boyd, Robert S. (November 11, 2002). "ANY BEING OUT THERE? Extreme Earth environments test astrobiology ideas". Philadelphia Inquirer. Retrieved 6 July 2016.
- ↑ Greenwood, Veronique (November 9, 2009). "What Life Leaves Behind What We Know: The search for life beyond our pale blue dot is fraught with dashed hopes. Will the chemical and mineral fingerprints of Earthly organisms apply on other worlds?". Seed Magazine. Archived from the original on November 15, 2009. Retrieved 6 July 2016.
{{cite journal}}: CS1 maint: unfit URL (link) - ↑ Benner, Steven A.; Hutter, Daniel (2002-02-01). "Phosphates, DNA, and the Search for Nonterrean Life: A Second Generation Model for Genetic Molecules". Bioorganic Chemistry. 30 (1): 62–80. doi:10.1006/bioo.2001.1232.
- ↑ Benner, Steven A. (2023-02-27). "Rethinking nucleic acids from their origins to their applications". Philosophical Transactions of the Royal Society B: Biological Sciences. 378 (1871). doi:10.1098/rstb.2022.0027. ISSN 0962-8436. PMC 9835595 .
- ↑ Špaček, Jan; Benner, Steven A. (2022-10-01). "Agnostic Life Finder (ALF) for Large-Scale Screening of Martian Life During In Situ Refueling". Astrobiology. 22 (10): 1255–1263. doi:10.1089/ast.2021.0070. ISSN 1531-1074.