
An electron microscope is a microscope that uses a beam of electrons as a source of illumination. They use electron optics that are analogous to the glass lenses of an optical light microscope to control the electron beam, for instance focusing them to produce magnified images or electron diffraction patterns. As the wavelength of an electron can be up to 100,000 times smaller than that of visible light, electron microscopes have a much higher resolution of about 0.1 nm, which compares to about 200 nm for light microscopes. Electron microscope may refer to:

Histology, also known as microscopic anatomy or microanatomy, is the branch of biology that studies the microscopic anatomy of biological tissues. Histology is the microscopic counterpart to gross anatomy, which looks at larger structures visible without a microscope. Although one may divide microscopic anatomy into organology, the study of organs, histology, the study of tissues, and cytology, the study of cells, modern usage places all of these topics under the field of histology. In medicine, histopathology is the branch of histology that includes the microscopic identification and study of diseased tissue. In the field of paleontology, the term paleohistology refers to the histology of fossil organisms.

Microscopy is the technical field of using microscopes to view objects and areas of objects that cannot be seen with the naked eye. There are three well-known branches of microscopy: optical, electron, and scanning probe microscopy, along with the emerging field of X-ray microscopy.

The optical microscope, also referred to as a light microscope, is a type of microscope that commonly uses visible light and a system of lenses to generate magnified images of small objects. Optical microscopes are the oldest design of microscope and were possibly invented in their present compound form in the 17th century. Basic optical microscopes can be very simple, although many complex designs aim to improve resolution and sample contrast.
An ultramicroscope is a microscope with a system that lights the object in a way that allows viewing of tiny particles via light scattering, and not light reflection or absorption. When the diameter of a particle is below or near the wavelength of visible light, the particle cannot be seen in a light microscope with the usual methods of illumination. The ultra- in ultramicroscope refers to the ability to see objects whose diameter is shorter than the wavelength of visible light, on the model of the ultra- in ultraviolet.

A fluorescence microscope is an optical microscope that uses fluorescence instead of, or in addition to, scattering, reflection, and attenuation or absorption, to study the properties of organic or inorganic substances. "Fluorescence microscope" refers to any microscope that uses fluorescence to generate an image, whether it is a simple set up like an epifluorescence microscope or a more complicated design such as a confocal microscope, which uses optical sectioning to get better resolution of the fluorescence image.

Confocal microscopy, most frequently confocal laser scanning microscopy (CLSM) or laser scanning confocal microscopy (LSCM), is an optical imaging technique for increasing optical resolution and contrast of a micrograph by means of using a spatial pinhole to block out-of-focus light in image formation. Capturing multiple two-dimensional images at different depths in a sample enables the reconstruction of three-dimensional structures within an object. This technique is used extensively in the scientific and industrial communities and typical applications are in life sciences, semiconductor inspection and materials science.

Two-photon excitation microscopy is a fluorescence imaging technique that is particularly well-suited to image scattering living tissue of up to about one millimeter in thickness. Unlike traditional fluorescence microscopy, where the excitation wavelength is shorter than the emission wavelength, two-photon excitation requires simultaneous excitation by two photons with longer wavelength than the emitted light. The laser is focused onto a specific location in the tissue and scanned across the sample to sequentially produce the image. Due to the non-linearity of two-photon excitation, mainly fluorophores in the micrometer-sized focus of the laser beam are excited, which results in the spatial resolution of the image. This contrasts with confocal microscopy, where the spatial resolution is produced by the interaction of excitation focus and the confined detection with a pinhole.

Phase-contrast microscopy (PCM) is an optical microscopy technique that converts phase shifts in light passing through a transparent specimen to brightness changes in the image. Phase shifts themselves are invisible, but become visible when shown as brightness variations.

Medical microbiology, the large subset of microbiology that is applied to medicine, is a branch of medical science concerned with the prevention, diagnosis and treatment of infectious diseases. In addition, this field of science studies various clinical applications of microbes for the improvement of health. There are four kinds of microorganisms that cause infectious disease: bacteria, fungi, parasites and viruses, and one type of infectious protein called prion.

Immunolabeling is a biochemical process that enables the detection and localization of an antigen to a particular site within a cell, tissue, or organ. Antigens are organic molecules, usually proteins, capable of binding to an antibody. These antigens can be visualized using a combination of antigen-specific antibody as well as a means of detection, called a tag, that is covalently linked to the antibody. If the immunolabeling process is meant to reveal information about a cell or its substructures, the process is called immunocytochemistry. Immunolabeling of larger structures is called immunohistochemistry.

Intravital microscopy is a form of microscopy that allows observing biological processes in live animals at a high resolution that makes distinguishing between individual cells of a tissue possible.

Cytometry is the measurement of number and characteristics of cells. Variables that can be measured by cytometric methods include cell size, cell count, cell morphology, cell cycle phase, DNA content, and the existence or absence of specific proteins on the cell surface or in the cytoplasm. Cytometry is used to characterize and count blood cells in common blood tests such as the complete blood count. In a similar fashion, cytometry is also used in cell biology research and in medical diagnostics to characterize cells in a wide range of applications associated with diseases such as cancer and AIDS.
Super-resolution microscopy is a series of techniques in optical microscopy that allow such images to have resolutions higher than those imposed by the diffraction limit, which is due to the diffraction of light. Super-resolution imaging techniques rely on the near-field or on the far-field. Among techniques that rely on the latter are those that improve the resolution only modestly beyond the diffraction-limit, such as confocal microscopy with closed pinhole or aided by computational methods such as deconvolution or detector-based pixel reassignment, the 4Pi microscope, and structured-illumination microscopy technologies such as SIM and SMI.
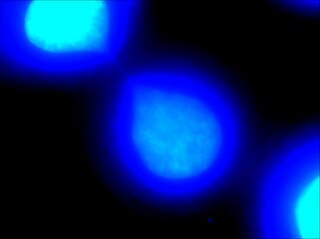
Chromatin bridge is a mitotic occurrence that forms when telomeres of sister chromatids fuse together and fail to completely segregate into their respective daughter cells. Because this event is most prevalent during anaphase, the term anaphase bridge is often used as a substitute. After the formation of individual daughter cells, the DNA bridge connecting homologous chromosomes remains fixed. As the daughter cells exit mitosis and re-enter interphase, the chromatin bridge becomes known as an interphase bridge. These phenomena are usually visualized using the laboratory techniques of staining and fluorescence microscopy.

Quantitative phase contrast microscopy or quantitative phase imaging are the collective names for a group of microscopy methods that quantify the phase shift that occurs when light waves pass through a more optically dense object.
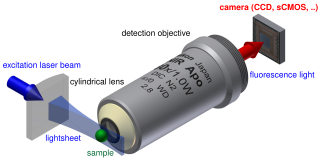
Light sheet fluorescence microscopy (LSFM) is a fluorescence microscopy technique with an intermediate-to-high optical resolution, but good optical sectioning capabilities and high speed. In contrast to epifluorescence microscopy only a thin slice of the sample is illuminated perpendicularly to the direction of observation. For illumination, a laser light-sheet is used, i.e. a laser beam which is focused only in one direction. A second method uses a circular beam scanned in one direction to create the lightsheet. As only the actually observed section is illuminated, this method reduces the photodamage and stress induced on a living sample. Also the good optical sectioning capability reduces the background signal and thus creates images with higher contrast, comparable to confocal microscopy. Because light sheet fluorescence microscopy scans samples by using a plane of light instead of a point, it can acquire images at speeds 100 to 1,000 times faster than those offered by point-scanning methods.

Live-cell imaging is the study of living cells using time-lapse microscopy. It is used by scientists to obtain a better understanding of biological function through the study of cellular dynamics. Live-cell imaging was pioneered in the first decade of the 21st century. One of the first time-lapse microcinematographic films of cells ever made was made by Julius Ries, showing the fertilization and development of the sea urchin egg. Since then, several microscopy methods have been developed to study living cells in greater detail with less effort. A newer type of imaging using quantum dots have been used, as they are shown to be more stable. The development of holotomographic microscopy has disregarded phototoxicity and other staining-derived disadvantages by implementing digital staining based on cells’ refractive index.

Cell unroofing is any of various methods to isolate and expose the cell membrane of cells. Differently from the more common membrane extraction protocols performed with multiple steps of centrifugation, in cell unroofing the aim is to tear and preserve patches of the plasma membrane in order to perform in situ experiments using.
Jean Comandon was a French microbiologist and filmmaker. He was one of the leading figures in the development of microcinematography in Paris and its use in science research and education.





















