
In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part of biological inheritance. This is essential for cell division during growth and repair of damaged tissues, while it also ensures that each of the new cells receives its own copy of the DNA. The cell possesses the distinctive property of division, which makes replication of DNA essential.

Transcription is the process of copying a segment of DNA into RNA. The segments of DNA transcribed into RNA molecules that can encode proteins produce messenger RNA (mRNA). Other segments of DNA are transcribed into RNA molecules called non-coding RNAs (ncRNAs).

In molecular biology, RNA polymerase, or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that catalyzes the chemical reactions that synthesize RNA from a DNA template.

A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create two identical DNA duplexes from a single original DNA duplex. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones. These enzymes catalyze the chemical reaction

DNA synthesis is the natural or artificial creation of deoxyribonucleic acid (DNA) molecules. DNA is a macromolecule made up of nucleotide units, which are linked by covalent bonds and hydrogen bonds, in a repeating structure. DNA synthesis occurs when these nucleotide units are joined to form DNA; this can occur artificially or naturally. Nucleotide units are made up of a nitrogenous base, pentose sugar (deoxyribose) and phosphate group. Each unit is joined when a covalent bond forms between its phosphate group and the pentose sugar of the next nucleotide, forming a sugar-phosphate backbone. DNA is a complementary, double stranded structure as specific base pairing occurs naturally when hydrogen bonds form between the nucleotide bases.
A sigma factor is a protein needed for initiation of transcription in bacteria. It is a bacterial transcription initiation factor that enables specific binding of RNA polymerase (RNAP) to gene promoters. It is homologous to archaeal transcription factor B and to eukaryotic factor TFIIB. The specific sigma factor used to initiate transcription of a given gene will vary, depending on the gene and on the environmental signals needed to initiate transcription of that gene. Selection of promoters by RNA polymerase is dependent on the sigma factor that associates with it. They are also found in plant chloroplasts as a part of the bacteria-like plastid-encoded polymerase (PEP).
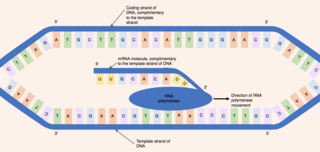
When referring to DNA transcription, the coding strand is the DNA strand whose base sequence is identical to the base sequence of the RNA transcript produced. It is this strand which contains codons, while the non-coding strand contains anticodons. During transcription, RNA Pol II binds to the non-coding template strand, reads the anti-codons, and transcribes their sequence to synthesize an RNA transcript with complementary bases.

The preinitiation complex is a complex of approximately 100 proteins that is necessary for the transcription of protein-coding genes in eukaryotes and archaea. The preinitiation complex positions RNA polymerase II at gene transcription start sites, denatures the DNA, and positions the DNA in the RNA polymerase II active site for transcription.

A primary transcript is the single-stranded ribonucleic acid (RNA) product synthesized by transcription of DNA, and processed to yield various mature RNA products such as mRNAs, tRNAs, and rRNAs. The primary transcripts designated to be mRNAs are modified in preparation for translation. For example, a precursor mRNA (pre-mRNA) is a type of primary transcript that becomes a messenger RNA (mRNA) after processing.

RNA polymerase II is a multiprotein complex that transcribes DNA into precursors of messenger RNA (mRNA) and most small nuclear RNA (snRNA) and microRNA. It is one of the three RNAP enzymes found in the nucleus of eukaryotic cells. A 550 kDa complex of 12 subunits, RNAP II is the most studied type of RNA polymerase. A wide range of transcription factors are required for it to bind to upstream gene promoters and begin transcription.

General transcription factors (GTFs), also known as basal transcriptional factors, are a class of protein transcription factors that bind to specific sites (promoter) on DNA to activate transcription of genetic information from DNA to messenger RNA. GTFs, RNA polymerase, and the mediator constitute the basic transcriptional apparatus that first bind to the promoter, then start transcription. GTFs are also intimately involved in the process of gene regulation, and most are required for life.
In eukaryote cells, RNA polymerase III is a protein that transcribes DNA to synthesize 5S ribosomal RNA, tRNA, and other small RNAs.

The replisome is a complex molecular machine that carries out replication of DNA. The replisome first unwinds double stranded DNA into two single strands. For each of the resulting single strands, a new complementary sequence of DNA is synthesized. The total result is formation of two new double stranded DNA sequences that are exact copies of the original double stranded DNA sequence.

T7 RNA Polymerase is an RNA polymerase from the T7 bacteriophage that catalyzes the formation of RNA from DNA in the 5'→ 3' direction.

Bacterial transcription is the process in which a segment of bacterial DNA is copied into a newly synthesized strand of messenger RNA (mRNA) with use of the enzyme RNA polymerase.

Eukaryotic transcription is the elaborate process that eukaryotic cells use to copy genetic information stored in DNA into units of transportable complementary RNA replica. Gene transcription occurs in both eukaryotic and prokaryotic cells. Unlike prokaryotic RNA polymerase that initiates the transcription of all different types of RNA, RNA polymerase in eukaryotes comes in three variations, each translating a different type of gene. A eukaryotic cell has a nucleus that separates the processes of transcription and translation. Eukaryotic transcription occurs within the nucleus where DNA is packaged into nucleosomes and higher order chromatin structures. The complexity of the eukaryotic genome necessitates a great variety and complexity of gene expression control.
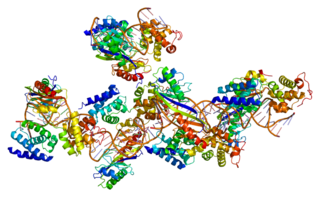
Transcription factor II B (TFIIB) is a general transcription factor that is involved in the formation of the RNA polymerase II preinitiation complex (PIC) and aids in stimulating transcription initiation. TFIIB is localised to the nucleus and provides a platform for PIC formation by binding and stabilising the DNA-TBP complex and by recruiting RNA polymerase II and other transcription factors. It is encoded by the TFIIB gene, and is homologous to archaeal transcription factor B and analogous to bacterial sigma factors.
RNA polymerase II holoenzyme is a form of eukaryotic RNA polymerase II that is recruited to the promoters of protein-coding genes in living cells. It consists of RNA polymerase II, a subset of general transcription factors, and regulatory proteins known as SRB proteins.
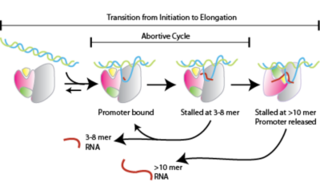
Abortive initiation, also known as abortive transcription, is an early process of genetic transcription in which RNA polymerase binds to a DNA promoter and enters into cycles of synthesis of short mRNA transcripts which are released before the transcription complex leaves the promoter. This process occurs in both eukaryotes and prokaryotes. Abortive initiation is typically studied in the T3 and T7 RNA polymerases in bacteriophages and in E. coli.

Archaeal transcription is the process in which a segment of archaeal DNA is copied into a newly synthesized strand of RNA using the sole Pol II-like RNA polymerase (RNAP). The process occurs in three main steps: initiation, elongation, and termination; and the end result is a strand of RNA that is complementary to a single strand of DNA. A number of transcription factors govern this process with homologs in both bacteria and eukaryotes, with the core machinery more similar to eukaryotic transcription.














