Related Research Articles

Evolution is the change in the heritable characteristics of biological populations over successive generations. Evolution occurs when evolutionary processes such as natural selection and genetic drift act on genetic variation, resulting in certain characteristics becoming more or less common within a population over successive generations. The process of evolution has given rise to biodiversity at every level of biological organisation.

In genetics, the phenotype is the set of observable characteristics or traits of an organism. The term covers the organism's morphology, its developmental processes, its biochemical and physiological properties, its behavior, and the products of behavior. An organism's phenotype results from two basic factors: the expression of an organism's genetic code and the influence of environmental factors. Both factors may interact, further affecting the phenotype. When two or more clearly different phenotypes exist in the same population of a species, the species is called polymorphic. A well-documented example of polymorphism is Labrador Retriever coloring; while the coat color depends on many genes, it is clearly seen in the environment as yellow, black, and brown. Richard Dawkins in 1978 and then again in his 1982 book The Extended Phenotype suggested that one can regard bird nests and other built structures such as caddisfly larva cases and beaver dams as "extended phenotypes".
Molecular evolution is the process of change in the sequence composition of cellular molecules such as DNA, RNA, and proteins across generations. The field of molecular evolution uses principles of evolutionary biology and population genetics to explain patterns in these changes. Major topics in molecular evolution concern the rates and impacts of single nucleotide changes, neutral evolution vs. natural selection, origins of new genes, the genetic nature of complex traits, the genetic basis of speciation, the evolution of development, and ways that evolutionary forces influence genomic and phenotypic changes.

Evolutionary biology is the subfield of biology that studies the evolutionary processes that produced the diversity of life on Earth. It is also defined as the study of the history of life forms on Earth. Evolution holds that all species are related and gradually change over generations. In a population, the genetic variations affect the phenotypes of an organism. These changes in the phenotypes will be an advantage to some organisms, which will then be passed on to their offspring. Some examples of evolution in species over many generations are the peppered moth and flightless birds. In the 1930s, the discipline of evolutionary biology emerged through what Julian Huxley called the modern synthesis of understanding, from previously unrelated fields of biological research, such as genetics and ecology, systematics, and paleontology.
Evolvability is defined as the capacity of a system for adaptive evolution. Evolvability is the ability of a population of organisms to not merely generate genetic diversity, but to generate adaptive genetic diversity, and thereby evolve through natural selection.
Genetic architecture is the underlying genetic basis of a phenotypic trait and its variational properties. Phenotypic variation for quantitative traits is, at the most basic level, the result of the segregation of alleles at quantitative trait loci (QTL). Environmental factors and other external influences can also play a role in phenotypic variation. Genetic architecture is a broad term that can be described for any given individual based on information regarding gene and allele number, the distribution of allelic and mutational effects, and patterns of pleiotropy, dominance, and epistasis.
The theory of facilitated variation demonstrates how seemingly complex biological systems can arise through a limited number of regulatory genetic changes, through the differential re-use of pre-existing developmental components. The theory was presented in 2005 by Marc W. Kirschner and John C. Gerhart.
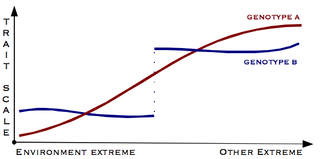
Canalisation is a measure of the ability of a population to produce the same phenotype regardless of variability of its environment or genotype. It is a form of evolutionary robustness. The term was coined in 1942 by C. H. Waddington to capture the fact that "developmental reactions, as they occur in organisms submitted to natural selection...are adjusted so as to bring about one definite end-result regardless of minor variations in conditions during the course of the reaction". He used this word rather than robustness to consider that biological systems are not robust in quite the same way as, for example, engineered systems.

A nuclear gene is a gene that has its DNA nucleotide sequence physically situated within the cell nucleus of a eukaryotic organism. This term is employed to differentiate nuclear genes, which are located in the cell nucleus, from genes that are found in mitochondria or chloroplasts. The vast majority of genes in eukaryotes are nuclear.
Biological constraints are factors which make populations resistant to evolutionary change. One proposed definition of constraint is "A property of a trait that, although possibly adaptive in the environment in which it originally evolved, acts to place limits on the production of new phenotypic variants." Constraint has played an important role in the development of such ideas as homology and body plans.

Evolutionary physiology is the study of the biological evolution of physiological structures and processes; that is, the manner in which the functional characteristics of individuals in a population of organisms have responded to natural selection across multiple generations during the history of the population. It is a sub-discipline of both physiology and evolutionary biology. Practitioners in the field come from a variety of backgrounds, including physiology, evolutionary biology, ecology, and genetics.
The following outline is provided as an overview of and topical guide to genetics:

In evolutionary biology, robustness of a biological system is the persistence of a certain characteristic or trait in a system under perturbations or conditions of uncertainty. Robustness in development is known as canalization. According to the kind of perturbation involved, robustness can be classified as mutational, environmental, recombinational, or behavioral robustness etc. Robustness is achieved through the combination of many genetic and molecular mechanisms and can evolve by either direct or indirect selection. Several model systems have been developed to experimentally study robustness and its evolutionary consequences.

Phenotypic Integration is a metric for measuring the correlation of multiple functionally-related traits to each other. Complex phenotypes often require multiple traits working together in order to function properly. Phenotypic integration is significant because it provides an explanation as to how phenotypes are sustained by relationships between traits. Every organism's phenotype is integrated, organized, and a functional whole. Integration is also associated with functional modules. Modules are complex character units that are tightly associated, such as a flower. It is hypothesized that organisms with high correlations between traits in a module have the most efficient functions. The fitness of a particular value for one phenotypic trait frequently depends on the value of the other phenotypic traits, making it important for those traits evolve together. One trait can have a direct effect on fitness, and it has been shown that the correlations among traits can also change fitness, causing these correlations to be adaptive, rather than solely genetic. Integration can be involved in multiple aspects of life, not just at the genetic level, but during development, or simply at a functional level.

The following outline is provided as an overview of and topical guide to evolution:
The Extended Evolutionary Synthesis (EES) consists of a set of theoretical concepts argued to be more comprehensive than the earlier modern synthesis of evolutionary biology that took place between 1918 and 1942. The extended evolutionary synthesis was called for in the 1950s by C. H. Waddington, argued for on the basis of punctuated equilibrium by Stephen Jay Gould and Niles Eldredge in the 1980s, and was reconceptualized in 2007 by Massimo Pigliucci and Gerd B. Müller.
In biology, constructive development refers to the hypothesis that organisms shape their own developmental trajectory by constantly responding to, and causing, changes in both their internal state and their external environment. Constructive development can be contrasted with programmed development, the hypothesis that organisms develop according to a genetic program or blueprint. The constructivist perspective is found in philosophy, most notably developmental systems theory, and in the biological and social sciences, including developmental psychobiology and key themes of the extended evolutionary synthesis. Constructive development may be important to evolution because it enables organisms to produce functional phenotypes in response to genetic or environmental perturbation, and thereby contributes to adaptation and diversification.
This glossary of genetics and evolutionary biology is a list of definitions of terms and concepts used in the study of genetics and evolutionary biology, as well as sub-disciplines and related fields, with an emphasis on classical genetics, quantitative genetics, population biology, phylogenetics, speciation, and systematics. Overlapping and related terms can be found in Glossary of cellular and molecular biology, Glossary of ecology, and Glossary of biology.
In evolutionary biology, developmental bias refers to the production against or towards certain ontogenetic trajectories which ultimately influence the direction and outcome of evolutionary change by affecting the rates, magnitudes, directions and limits of trait evolution. Historically, the term was synonymous with developmental constraint, however, the latter has been more recently interpreted as referring solely to the negative role of development in evolution.
Lee Altenberg is an American theoretical biologist. He is on the faculty of the Departments of Information and Computer Sciences and of Mathematics at the University of Hawaiʻi at Mānoa. He is best known for his work that helped establish the evolution of evolvability and modularity in the genotype–phenotype map as areas of investigation in evolutionary biology, for moving theoretical concepts between the fields of evolutionary biology and evolutionary computation, and for his mathematical unification and generalization of modifier gene models for the evolution of biological information transmission, putting under a single mathematical framework the evolution of mutation rates, recombination rates, sexual reproduction rates, and dispersal rates.
References
- ↑ Altenberg, L. 1994. The evolution of evolvability in genetic programming. Chapter 3 in Advances in Genetic Programming, ed. Kenneth Kinnear. MIT Press, pp. 47-74.
- ↑ Altenberg, L. 1995. Genome Growth and the Evolution of the Genotype-Phenotype Map In Evolution and Biocomputation: Computational Models of Evolution. Wolfgang Banzhaf and Frank H. Eeckman (Eds.). pp. 205-259. Berlin: Springer.
- ↑ Wagner, G. P. and Altenberg, L. 1996. Complex adaptations and the evolution of evolvability. Evolution 50 (3): 967-976.