Molecular biology is the branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions. The study of chemical and physical structure of biological macromolecules is known as molecular biology.

Nucleic acids are biopolymers, macromolecules, essential to all known forms of life. They are composed of nucleotides, which are the monomer components: a 5-carbon sugar, a phosphate group and a nitrogenous base. The two main classes of nucleic acids are deoxyribonucleic acid (DNA) and ribonucleic acid (RNA). If the sugar is ribose, the polymer is RNA; if the sugar is the ribose derivative deoxyribose, the polymer is DNA.
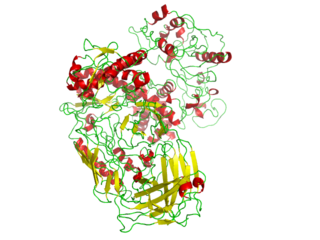
A polymerase is an enzyme that synthesizes long chains of polymers or nucleic acids. DNA polymerase and RNA polymerase are used to assemble DNA and RNA molecules, respectively, by copying a DNA template strand using base-pairing interactions or RNA by half ladder replication.

Ribosomes are macromolecular machines, found within all cells, that perform biological protein synthesis. Ribosomes link amino acids together in the order specified by the codons of messenger RNA (mRNA) molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA (rRNA) molecules and many ribosomal proteins. The ribosomes and associated molecules are also known as the translational apparatus.

The central dogma of molecular biology is an explanation of the flow of genetic information within a biological system. It is often stated as "DNA makes RNA, and RNA makes protein", although this is not its original meaning. It was first stated by Francis Crick in 1957, then published in 1958:
The Central Dogma. This states that once "information" has passed into protein it cannot get out again. In more detail, the transfer of information from nucleic acid to nucleic acid, or from nucleic acid to protein may be possible, but transfer from protein to protein, or from protein to nucleic acid is impossible. Information here means the precise determination of sequence, either of bases in the nucleic acid or of amino acid residues in the protein.

In molecular biology, RNA polymerase, or more specifically DNA-directed/dependent RNA polymerase (DdRP), is an enzyme that catalyzes the chemical reactions that synthesize RNA from a DNA template.

A primary transcript is the single-stranded ribonucleic acid (RNA) product synthesized by transcription of DNA, and processed to yield various mature RNA products such as mRNAs, tRNAs, and rRNAs. The primary transcripts designated to be mRNAs are modified in preparation for translation. For example, a precursor mRNA (pre-mRNA) is a type of primary transcript that becomes a messenger RNA (mRNA) after processing.
The Shine–Dalgarno (SD) sequence is a ribosomal binding site in bacterial and archaeal messenger RNA, generally located around 8 bases upstream of the start codon AUG. The RNA sequence helps recruit the ribosome to the messenger RNA (mRNA) to initiate protein synthesis by aligning the ribosome with the start codon. Once recruited, tRNA may add amino acids in sequence as dictated by the codons, moving downstream from the translational start site.

The Max Planck Institute of Biochemistry (MPIB) is a research institute of the Max Planck Society located in Martinsried, a suburb of Munich. The institute was founded in 1973 by the merger of three formerly independent institutes: the Max Planck Institute of Biochemistry, the Max Planck Institute of Protein and Leather Research, and the Max Planck Institute of Cell Chemistry.
The history of molecular biology begins in the 1930s with the convergence of various, previously distinct biological and physical disciplines: biochemistry, genetics, microbiology, virology and physics. With the hope of understanding life at its most fundamental level, numerous physicists and chemists also took an interest in what would become molecular biology.

In biology, the word gene can have several different meanings. The Mendelian gene is a basic unit of heredity and the molecular gene is a sequence of nucleotides in DNA that is transcribed to produce a functional RNA. There are two types of molecular genes: protein-coding genes and noncoding genes.

In molecular biology, the 5.8S ribosomal RNA is a non-coding RNA component of the large subunit of the eukaryotic ribosome and so plays an important role in protein translation. It is transcribed by RNA polymerase I as part of the 45S precursor that also contains 18S and 28S rRNA. Its function is thought to be in ribosome translocation. It is also known to form covalent linkage to the p53 tumour suppressor protein. 5.8S rRNA can be used as a reference gene for miRNA detection. The 5.8S ribosomal RNA is used to better understand other rRNA processes and pathways in the cell.

In molecular biology, Small nucleolar RNA U6-53 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, Small nucleolar RNA psi28S-3327 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a 'guide RNA'.
In molecular biology, SNORA4 is a member of the H/ACA class of small nucleolar RNA that guide the sites of modification of uridines to pseudouridines.
In molecular biology, Small nucleolar RNA SNORA48 is a pseudouridylation guide H/ACA box snoRNA. This snoRNA was cloned in 2004 from a HeLa cell extract immunoprecipitated with an anti-GAR1 antibody. It is predicted to guide the pseudouridylation of residue U3797 of 28S rRNA.

In molecular biology, Small nucleolar RNA SNORA74 (U19) belongs to the H/ACA class of snoRNAs. snoRNAs bind a number of proteins to form snoRNP complexes. This class is thought to guide the sites of modification of uridines to pseudouridines by forming direct base pairing interactions with substrate RNAs. Targets may include ribosomal and spliceosomal RNAs but the exact functions of many snoRNAs, including U19, are not confirmed. Co-precipitation of U19 snoRNA with RNase MRP RNA suggests that U19 may be involved in pre-rRNA processing.

In molecular biology, snoRNA U38 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, Small nucleolar RNA SNORD83 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA SNORD83 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. snoRNA SNORD83 are spliced from introns 5 and 4 of the BAT1 gene in mammals.
In molecular biology, snoRNA Z17 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

















