
Glucocorticoids are a class of corticosteroids, which are a class of steroid hormones. Glucocorticoids are corticosteroids that bind to the glucocorticoid receptor that is present in almost every vertebrate animal cell. The name "glucocorticoid" is a portmanteau and is composed from its role in regulation of glucose metabolism, synthesis in the adrenal cortex, and its steroidal structure.
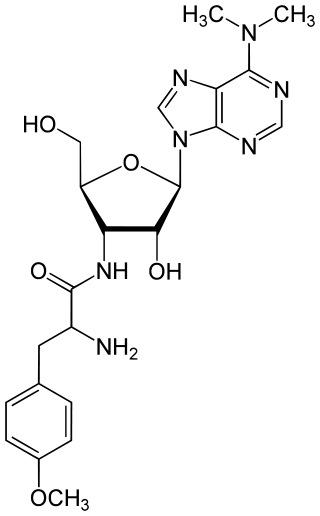
Puromycin is an antibiotic protein synthesis inhibitor which causes premature chain termination during translation.

Anisomycin, also known as flagecidin, is an antibiotic produced by Streptomyces griseolus which inhibits eukaryotic protein synthesis. Partial inhibition of DNA synthesis occurs at anisomycin concentrations that effect 95% inhibition of protein synthesis. Anisomycin can activate stress-activated protein kinases, MAP kinase and other signal transduction pathways.
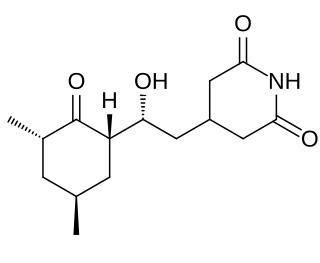
Cycloheximide is a naturally occurring fungicide produced by the bacterium Streptomyces griseus. Cycloheximide exerts its effects by interfering with the translocation step in protein synthesis, thus blocking eukaryotic translational elongation. Cycloheximide is widely used in biomedical research to inhibit protein synthesis in eukaryotic cells studied in vitro. It is inexpensive and works rapidly. Its effects are rapidly reversed by simply removing it from the culture medium.

Filamentation is the anomalous growth of certain bacteria, such as Escherichia coli, in which cells continue to elongate but do not divide. The cells that result from elongation without division have multiple chromosomal copies.
A bacterivore is an organism which obtains energy and nutrients primarily or entirely from the consumption of bacteria. The term is most commonly used to describe free-living, heterotrophic, microscopic organisms such as nematodes as well as many species of amoeba and numerous other types of protozoans, but some macroscopic invertebrates are also bacterivores, including sponges, polychaetes, and certain molluscs and arthropods. Many bacterivorous organisms are adapted for generalist predation on any species of bacteria, but not all bacteria are easily digested; the spores of some species, such as Clostridium perfringens, will never be prey because of their cellular attributes.

Monoamine oxidase B, also known as MAOB, is an enzyme that in humans is encoded by the MAOB gene.

60S ribosomal protein L7a is a protein that in humans is encoded by the RPL7A gene.
Calcineurin-binding protein cabin-1 is a protein that in humans is encoded by the CABIN1 gene.

Eukaryotic translation initiation factor 2A (eIF2A) is a protein that in humans is encoded by the EIF2A gene. The eIF2A protein is not to be confused with eIF2α, a subunit of the heterotrimeric eIF2 complex. Instead, eIF2A functions by a separate mechanism in eukaryotic translation.
Memory consolidation is a category of processes that stabilize a memory trace after its initial acquisition. A memory trace is a change in the nervous system caused by memorizing something. Consolidation is distinguished into two specific processes. The first, synaptic consolidation, which is thought to correspond to late-phase long-term potentiation, occurs on a small scale in the synaptic connections and neural circuits within the first few hours after learning. The second process is systems consolidation, occurring on a much larger scale in the brain, rendering hippocampus-dependent memories independent of the hippocampus over a period of weeks to years. Recently, a third process has become the focus of research, reconsolidation, in which previously consolidated memories can be made labile again through reactivation of the memory trace.
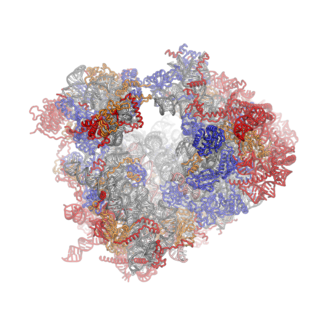
Ribosomes are a large and complex molecular machine that catalyzes the synthesis of proteins, referred to as translation. The ribosome selects aminoacylated transfer RNAs (tRNAs) based on the sequence of a protein-encoding messenger RNA (mRNA) and covalently links the amino acids into a polypeptide chain. Ribosomes from all organisms share a highly conserved catalytic center. However, the ribosomes of eukaryotes are much larger than prokaryotic ribosomes and subject to more complex regulation and biogenesis pathways. Eukaryotic ribosomes are also known as 80S ribosomes, referring to their sedimentation coefficients in Svedberg units, because they sediment faster than the prokaryotic (70S) ribosomes. Eukaryotic ribosomes have two unequal subunits, designated small subunit (40S) and large subunit (60S) according to their sedimentation coefficients. Both subunits contain dozens of ribosomal proteins arranged on a scaffold composed of ribosomal RNA (rRNA). The small subunit monitors the complementarity between tRNA anticodon and mRNA, while the large subunit catalyzes peptide bond formation.

Dynamic combinatorial chemistry (DCC); also known as constitutional dynamic chemistry (CDC) is a method to the generation of new molecules formed by reversible reaction of simple building blocks under thermodynamic control. The library of these reversibly interconverting building blocks is called a dynamic combinatorial library (DCL). All constituents in a DCL are in equilibrium, and their distribution is determined by their thermodynamic stability within the DCL. The interconversion of these building blocks may involve covalent or non-covalent interactions. When a DCL is exposed to an external influence, the equilibrium shifts and those components that interact with the external influence are stabilised and amplified, allowing more of the active compound to be formed.

Glutarimide is the organic compound with the formula (CH2)3(CO)2NH. It is a white solid. The compound forms upon dehydration of the amide of glutaric acid.

Ribosome profiling, or Ribo-Seq, is an adaptation of a technique developed by Joan Steitz and Marilyn Kozak almost 50 years ago that Nicholas Ingolia and Jonathan Weissman adapted to work with next generation sequencing that uses specialized messenger RNA (mRNA) sequencing to determine which mRNAs are being actively translated. A related technique that can also be used to determine which mRNAs are being actively translated is the Translating Ribosome Affinity Purification (TRAP) methodology, which was developed by Nathaniel Heintz at Rockefeller University. TRAP does not involve ribosome footprinting but provides cell type-specific information.
The de novo protein synthesis theory of memory formation is a hypothesis about the formation of the physical correlates of memory in the brain. It is widely accepted that the physiological correlates for memories are stored at the synapse between various neurons. The relative strength of various synapses in a network of neurons form the memory trace, or ‘engram,’ though the processes that support this finding are less thoroughly understood. The de novo protein synthesis theory states that the production of proteins is required to initiate and potentially maintain these plastic changes within the brain. It has much support within the neuroscience community, but some critics claim that memories can be made independent of protein synthesis.

mTORC1, also known as mammalian target of rapamycin complex 1 or mechanistic target of rapamycin complex 1, is a protein complex that functions as a nutrient/energy/redox sensor and controls protein synthesis.

Ribosomal pause refers to the queueing or stacking of ribosomes during translation of the nucleotide sequence of mRNA transcripts. These transcripts are decoded and converted into an amino acid sequence during protein synthesis by ribosomes. Due to the pause sites of some mRNA's, there is a disturbance caused in translation. Ribosomal pausing occurs in both eukaryotes and prokaryotes. A more severe pause is known as a ribosomal stall.
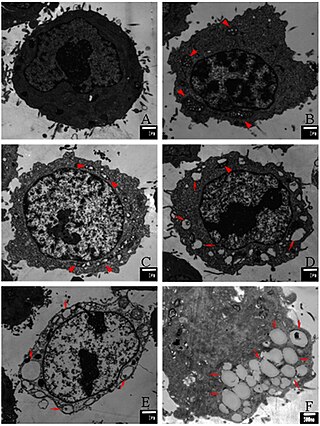
Paraptosis is a type of programmed cell death, morphologically distinct from apoptosis and necrosis. The defining features of paraptosis are cytoplasmic vacuolation, independent of caspase activation and inhibition, and lack of apoptotic morphology. Paraptosis lacks several of the hallmark characteristics of apoptosis, such as membrane blebbing, chromatin condensation, and nuclear fragmentation. Like apoptosis and other types of programmed cell death, the cell is involved in causing its own death, and gene expression is required. This is in contrast to necrosis, which is non-programmed cell death that results from injury to the cell.
Cycloheximide chase assays are an experimental technique used in molecular and cellular biology to measure steady state protein stability. Cycloheximide is a drug that inhibits the elongation step in eukaryotic protein translation, thereby preventing protein synthesis. The addition of cycloheximide to cultured cells followed by protein lysis at multiple timepoints is conducted to observe protein degradation over time and can be used to determine a protein's half-life. These assays are often followed by western blotting to assess protein abundance and can be analyzed using quantitative tools such as ImageJ.
















