In information science, an ontology encompasses a representation, formal naming, and definition of the categories, properties, and relations between the concepts, data, and entities that substantiate one, many, or all domains of discourse. More simply, an ontology is a way of showing the properties of a subject area and how they are related, by defining a set of concepts and categories that represent the subject.

Health informatics is the field of science and engineering that aims at developing methods and technologies for the acquisition, processing, and study of patient data, which can come from different sources and modalities, such as electronic health records, diagnostic test results, medical scans. The health domain provides an extremely wide variety of problems that can be tackled using computational techniques.
A medical classification is used to transform descriptions of medical diagnoses or procedures into standardized statistical code in a process known as clinical coding. Diagnosis classifications list diagnosis codes, which are used to track diseases and other health conditions, inclusive of chronic diseases such as diabetes mellitus and heart disease, and infectious diseases such as norovirus, the flu, and athlete's foot. Procedure classifications list procedure code, which are used to capture interventional data. These diagnosis and procedure codes are used by health care providers, government health programs, private health insurance companies, workers' compensation carriers, software developers, and others for a variety of applications in medicine, public health and medical informatics, including:
OpenGALEN is a not-for-profit organisation that provides an open source medical terminology. This terminology is written in a formal language called GRAIL and also distributed in OWL.
A clinical terminology server is a terminology server, which contains and provides access to clinical terminology.
The Unified Medical Language System (UMLS) is a compendium of many controlled vocabularies in the biomedical sciences. It provides a mapping structure among these vocabularies and thus allows one to translate among the various terminology systems; it may also be viewed as a comprehensive thesaurus and ontology of biomedical concepts. UMLS further provides facilities for natural language processing. It is intended to be used mainly by developers of systems in medical informatics.
The Systematized Nomenclature of Medicine (SNOMED) is a systematic, computer-processable collection of medical terms, in human and veterinary medicine, to provide codes, terms, synonyms and definitions which cover anatomy, diseases, findings, procedures, microorganisms, substances, etc. It allows a consistent way to index, store, retrieve, and aggregate medical data across specialties and sites of care. Although now international, SNOMED was started in the U.S. by the College of American Pathologists (CAP) in 1973 and revised into the 1990s. In 2002 CAP's SNOMED Reference Terminology was merged with, and expanded by, the National Health Service's Clinical Terms Version 3 to produce SNOMED CT.
Biomedical text mining refers to the methods and study of how text mining may be applied to texts and literature of the biomedical domain. As a field of research, biomedical text mining incorporates ideas from natural language processing, bioinformatics, medical informatics and computational linguistics. The strategies in this field have been applied to the biomedical literature available through services such as PubMed.

SNOMED CT or SNOMED Clinical Terms is a systematically organized computer-processable collection of medical terms providing codes, terms, synonyms and definitions used in clinical documentation and reporting. SNOMED CT is considered to be the most comprehensive, multilingual clinical healthcare terminology in the world. The primary purpose of SNOMED CT is to encode the meanings that are used in health information and to support the effective clinical recording of data with the aim of improving patient care. SNOMED CT provides the core general terminology for electronic health records. SNOMED CT comprehensive coverage includes: clinical findings, symptoms, diagnoses, procedures, body structures, organisms and other etiologies, substances, pharmaceuticals, devices and specimens.
CADUCEUS was a medical expert system - an early type of recommender system - by Harry Pople of the University of Pittsburgh. Finished in the mid-1980s, it was built on the INTERNIST-1 algorithm (1972-1973). In its time, CADUCEUS was described as the "most knowledge-intensive expert system in existence". CADUCEUS eventually could diagnose up to 1000 different diseases.
The Open Biological and Biomedical Ontologies (OBO) Foundry is a group of people dedicated to build and maintain ontologies related to the life sciences. The OBO Foundry establishes a set of principles for ontology development for creating a suite of interoperable reference ontologies in the biomedical domain. Currently, there are more than a hundred ontologies that follow the OBO Foundry principles.

Carole Anne Goble, is a British academic who is Professor of Computer Science at the University of Manchester. She is principal investigator (PI) of the myGrid, BioCatalogue and myExperiment projects and co-leads the Information Management Group (IMG) with Norman Paton.

Robert David Stevens is a professor of bio-health informatics. and former Head of Department of Computer Science at The University of Manchester
MEDCIN, a system of standardized medical terminology, is a proprietary medical vocabulary and was developed by Medicomp Systems, Inc. MEDCIN is a point-of-care terminology, intended for use in Electronic Health Record (EHR) systems, and it includes over 280,000 clinical data elements encompassing symptoms, history, physical examination, tests, diagnoses and therapy. This clinical vocabulary contains over 38 years of research and development as well as the capability to cross map to leading codification systems such as SNOMED CT, CPT, ICD-9-CM/ICD-10-CM, DSM, LOINC, CDT, CVX, and the Clinical Care Classification (CCC) System for nursing and allied health.

Frank van Harmelen is a Dutch computer scientist and professor in Knowledge Representation & Reasoning in the AI department at the Vrije Universiteit Amsterdam. He was scientific director of the LarKC project (2008-2011), "aiming to develop the Large Knowledge Collider, a platform for very large scale semantic web reasoning."

NeuroLex is a lexicon of neuroscience concepts supported by the Neuroscience Information Framework project. It is structured as a semantic wiki, using Semantic MediaWiki.
The Disease Ontology (DO) is a formal ontology of human disease. The Disease Ontology project is hosted at the Institute for Genome Sciences at the University of Maryland School of Medicine.
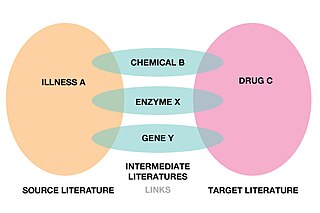
Literature-based discovery (LBD), also called literature-related discovery (LRD) is a form of knowledge extraction and automated hypothesis generation that uses papers and other academic publications to find new relationships between existing knowledge. Literature-based discovery aims to discover new knowledge by connecting information which have been explicitly stated in literature to deduce connections which have not been explicitly stated.

Dipak Kalra is President of the European Institute for Health Records and of the European Institute for Innovation through Health Data. He undertakes international research and standards development, and advises on adoption strategies, relating to Electronic Health Records.
Mark Alan Musen is a Professor of Biomedical Informatics and of Biomedical Data Science at Stanford University, and Division Director of the Stanford Center for Biomedical Informatics Research. Musen's research focuses on open science, data stewardship, intelligent systems, and biomedical decision support. Since the late 1980s, Musen has led the development of Protégé, which is currently the most "widely used domain-independent, freely available, platform-independent technology for developing and managing terminologies, ontologies, and knowledge bases" in a range of application domains.








