Related Research Articles

In mathematics, particularly in dynamical systems, a bifurcation diagram shows the values visited or approached asymptotically of a system as a function of a bifurcation parameter in the system. It is usual to represent stable values with a solid line and unstable values with a dotted line, although often the unstable points are omitted. Bifurcation diagrams enable the visualization of bifurcation theory. In the context of discrete-time dynamical systems, the diagram is also called orbit diagram.
In mathematics and science, a nonlinear system is a system in which the change of the output is not proportional to the change of the input. Nonlinear problems are of interest to engineers, biologists, physicists, mathematicians, and many other scientists since most systems are inherently nonlinear in nature. Nonlinear dynamical systems, describing changes in variables over time, may appear chaotic, unpredictable, or counterintuitive, contrasting with much simpler linear systems.
In signal processing and electronics, the frequency response of a system is the quantitative measure of the magnitude and phase of the output as a function of input frequency. The frequency response is widely used in the design and analysis of systems, such as audio and control systems, where they simplify mathematical analysis by converting governing differential equations into algebraic equations. In an audio system, it may be used to minimize audible distortion by designing components so that the overall response is as flat (uniform) as possible across the system's bandwidth. In control systems, such as a vehicle's cruise control, it may be used to assess system stability, often through the use of Bode plots. Systems with a specific frequency response can be designed using analog and digital filters.
Visual Molecular Dynamics (VMD) is a molecular modelling and visualization computer program. VMD is developed mainly as a tool to view and analyze the results of molecular dynamics simulations. It also includes tools for working with volumetric data, sequence data, and arbitrary graphics objects. Molecular scenes can be exported to external rendering tools such as POV-Ray, RenderMan, Tachyon, Virtual Reality Modeling Language (VRML), and many others. Users can run their own Tcl and Python scripts within VMD as it includes embedded Tcl and Python interpreters. VMD runs on Unix, Apple Mac macOS, and Microsoft Windows. VMD is available to non-commercial users under a distribution-specific license which permits both use of the program and modification of its source code, at no charge.

Physiologically based pharmacokinetic (PBPK) modeling is a mathematical modeling technique for predicting the absorption, distribution, metabolism and excretion (ADME) of synthetic or natural chemical substances in humans and other animal species. PBPK modeling is used in pharmaceutical research and drug development, and in health risk assessment for cosmetics or general chemicals.
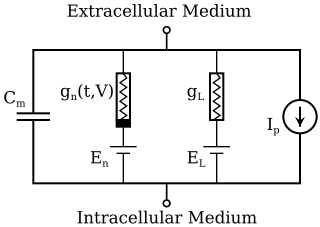
The Hodgkin–Huxley model, or conductance-based model, is a mathematical model that describes how action potentials in neurons are initiated and propagated. It is a set of nonlinear differential equations that approximates the electrical engineering characteristics of excitable cells such as neurons and muscle cells. It is a continuous-time dynamical system.

The dose–response relationship, or exposure–response relationship, describes the magnitude of the response of an organism, as a function of exposure to a stimulus or stressor after a certain exposure time. Dose–response relationships can be described by dose–response curves. This is explained further in the following sections. A stimulus response function or stimulus response curve is defined more broadly as the response from any type of stimulus, not limited to chemicals.
Systems immunology is a research field under systems biology that uses mathematical approaches and computational methods to examine the interactions within cellular and molecular networks of the immune system. The immune system has been thoroughly analyzed as regards to its components and function by using a "reductionist" approach, but its overall function can't be easily predicted by studying the characteristics of its isolated components because they strongly rely on the interactions among these numerous constituents. It focuses on in silico experiments rather than in vivo.
NONMEM is a non-linear mixed-effects modeling software package developed by Stuart L. Beal and Lewis B. Sheiner in the late 1970s at University of California, San Francisco, and expanded by Robert Bauer at Icon PLC. Its name is an acronym for NON-linear mixed effects modeling but it is especially powerful in the context of population pharmacokinetics, pharmacometrics, and PK/PD models. NONMEM models are written in NMTRAN, a dedicated model specification language that is translated into FORTRAN, compiled on the fly and executed by a command-line script. Results are presented as text output files including tables. There are multiple interfaces to assist modelers with housekeeping of files, tracking of model development, goodness-of-fit evaluations and graphical output, such as PsN and xpose and Wings for NONMEM. Current version for NONMEM is 7.5.
Pharmacometrics is a field of study of the methodology and application of models for disease and pharmacological measurement. It uses mathematical models of biology, pharmacology, disease, and physiology to describe and quantify interactions between xenobiotics and patients, including beneficial effects and adverse effects. It is normally applied to quantify drug, disease and trial information to aid efficient drug development, regulatory decisions and rational drug treatment in patients.

Denis Louis Blackmore was an American mathematician and a full professor of the Department of Mathematical Sciences at New Jersey Institute of Technology. He was also one of the founding members of the Center for Applied Mathematics and Statistics at NJIT. Dr. Blackmore was mainly known for his many contributions in the fields of dynamical systems and differential topology. In addition to this, he had many contributions in other fields of applied mathematics, physics, biology, and engineering.
In the field of pharmacokinetics, the area under the curve (AUC) is the definite integral of the concentration of a drug in blood plasma as a function of time. In practice, the drug concentration is measured at certain discrete points in time and the trapezoidal rule is used to estimate AUC. In pharmacology, the area under the plot of plasma concentration of a drug versus time after dosage gives insight into the extent of exposure to a drug and its clearance rate from the body.
PK/PD modeling is a technique that combines the two classical pharmacologic disciplines of pharmacokinetics and pharmacodynamics. It integrates a pharmacokinetic and a pharmacodynamic model component into one set of mathematical expressions that allows the description of the time course of effect intensity in response to administration of a drug dose. PK/PD modeling is related to the field of pharmacometrics.

Ronald J. DiPerna was an American mathematician, who worked on nonlinear partial differential equations.

John J. Tyson is an American systems biologist and mathematical biologist who serves as University Distinguished Professor of Biology at Virginia Tech, and is the former president of the Society for Mathematical Biology. He is known for his research on biochemical switches in the cell cycle, dynamics of biological networks and on excitable media.
Quantitative systems pharmacology (QSP) is a discipline within biomedical research that uses mathematical computer models to characterize biological systems, disease processes and drug pharmacology. QSP can be viewed as a sub-discipline of pharmacometrics that focuses on modeling the mechanisms of drug pharmacokinetics (PK), pharmacodynamics (PD), and disease processes using a systems pharmacology point of view. QSP models are typically defined by systems of ordinary differential equations (ODE) that depict the dynamical properties of the interaction between the drug and the biological system.
In mathematics and mathematical biology, the Mackey–Glass equations, named after Michael Mackey and Leon Glass, refer to a family of delay differential equations whose behaviour manages to mimic both healthy and pathological behaviour in certain biological contexts, controlled by the equation's parameters. Originally, they were used to model the variation in the relative quantity of mature cells in the blood. The equations are defined as:

SAAM II, short for "Simulation Analysis and Modeling" version 2.0, is a renowned computer program designed for scientific research in the field of bioscience. It is a descriptive and exploratory tool in drug development, tracers, metabolic disorders, and pharmacokinetics/pharmacodynamics research. It is grounded in the principles of multi-compartment model theory, which is a widely-used approach for modeling complex biological systems. SAAM II facilitates the construction and simulation of models, providing researchers with a friendly user interface allowing the quick run and multi-fitting of simple and complex structures and data. SAAM II is used by many Pharma and Pharmacy Schools as a drug development, research, and educational tool.
Target-mediated drug disposition (TMDD), is the process in which a drug binds with high affinity to its pharmacological target to such an extent that this affects its pharmacokinetic characteristics. Various drug classes can exhibit TMDD, most often these are large compounds but also smaller compounds can exhibit TMDD . A typical TMDD pattern of antibodies displays non-linear clearance and can be seen at concentration ranges that are usually defined as 'mid-to-low'. In this concentration range, the target is partly saturated.
Doron Levy is a mathematician, scientist, magician, and academic. He is a Professor and chair at the Department of Mathematics at the University of Maryland, College Park. He is also the Director of the Brin Mathematics Research Center.
References
- ↑ Macey, Robert; Oster. George; Zahnley, Tim (December 28, 2009). Berkeley Madonna User’s Guide Archived 2015-02-26 at the Wayback Machine University of California. Department of Molecular and Cellular Biology. Berkeley, California.
- ↑ Marcoline, Frank; Grabe, Michael; Nayak, Smita; Zahnley, Tim; Oster, George; Macey, Robert (February 28, 2021). Berkeley Madonna User’s Guide Version 10.2.6 Berkeley Madonna, Inc., Albany, CA 94706.
- ↑ Krause, A; Lowe, PJ (2014-05-28). "Visualization and Communication of Pharmacometric Models With Berkeley Madonna". CPT: Pharmacometrics & Systems Pharmacology. 3 (5): 113. doi:10.1038/psp.2014.13. PMC 4055786 . PMID 24872204. pp. 1–20.
- ↑ Marcoline, F; Furth, J; Nayak, S; Grabe, M; Macey, R.I. (2021-10-15). "Berkeley Madonna Version 10–A simulation package for solving mathematical models". CPT: Pharmacometrics & Systems Pharmacology. 11 (3): 290–301. doi:10.1002/psp4.12757. PMC 8923725 . PMID 35064965. pp. 290-301.
- ↑ Zhong, H.; Wade, S.M.; Woolf, P.J.; Linderman, J.J.; Traynor, J.R.; Neubig, R.R. (2003). "A Spatial Focusing Model for G Protein Signals". Journal of Biological Chemistry. 278 (9): 7278–7284. doi: 10.1074/jbc.m208819200 . PMID 12446706.
- ↑ "LinkedIn group Berkeley Madonna". LinkedIn.com. Retrieved February 8, 2023.
- ↑ Vinnycky, Emilia; White, Richard (2010-05-13). An introduction to infectious disease modelling. Oxford University Press. ISBN 978-0-19-856576-5.
- ↑ Robson, MG; Toscano, WA (2007). Risk Assessment for Environmental Health. John Wiley & Sons. ISBN 9780787988593.
- ↑ Weller, Florian; Sherley, Richard B.; Waller, Lauren J.; Ludynia, Katrin; Geldenhuys, Deon; Shannon, Lynne J.; Jarre, Astrid (2016). "System dynamics modelling of the Endangered African penguin populations on Dyer and Robben islands, South Africa". Ecological Modelling. 327: 44–56. Bibcode:2016EcMod.327...44W. doi:10.1016/j.ecolmodel.2016.01.011.