Related Research Articles

In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the most essential part of biological inheritance. This is essential for cell division during growth and repair of damaged tissues, while it also ensures that each of the new cells receives its own copy of the DNA. The cell possesses the distinctive property of division, which makes replication of DNA essential.

AstraZeneca plc (AZ) is a British-Swedish multinational pharmaceutical and biotechnology company with its headquarters at the Cambridge Biomedical Campus in Cambridge, England. It has a portfolio of products for major diseases in areas including oncology, cardiovascular, gastrointestinal, infection, neuroscience, respiratory, and inflammation. It has been involved in developing the Oxford–AstraZeneca COVID-19 vaccine.

Adenoviruses are medium-sized, nonenveloped viruses with an icosahedral nucleocapsid containing a double-stranded DNA genome. Their name derives from their initial isolation from human adenoids in 1953.

DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA damage, resulting in tens of thousands of individual molecular lesions per cell per day. Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes. Other lesions induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells after it undergoes mitosis. As a consequence, the DNA repair process is constantly active as it responds to damage in the DNA structure. When normal repair processes fail, and when cellular apoptosis does not occur, irreparable DNA damage may occur. This can eventually lead to malignant tumors, or cancer as per the two-hit hypothesis.
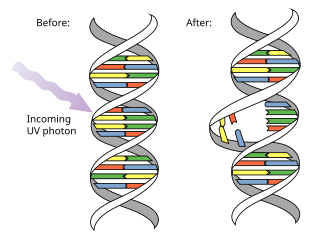
Pyrimidine dimers represent molecular lesions originating from thymine or cytosine bases within DNA, resulting from photochemical reactions. These lesions, commonly linked to direct DNA damage, are induced by ultraviolet light (UV), particularly UVC, result in the formation of covalent bonds between adjacent nitrogenous bases along the nucleotide chain near their carbon–carbon double bonds, the photo-coupled dimers are fluorescent. Such dimerization, which can also occur in double-stranded RNA (dsRNA) involving uracil or cytosine, leads to the creation of cyclobutane pyrimidine dimers (CPDs) and 6–4 photoproducts. These pre-mutagenic lesions modify the DNA helix structure, resulting in abnormal non-canonical base pairing and, consequently, adjacent thymines or cytosines in DNA will form a cyclobutane ring when joined together and cause a distortion in the DNA. This distortion prevents DNA replication and transcription mechanisms beyond the dimerization site.
Postreplication repair is the repair of damage to the DNA that takes place after replication.

Cell cycle checkpoint control protein RAD9A is a protein that in humans is encoded by the RAD9A gene.Rad9 has been shown to induce G2 arrest in the cell cycle in response to DNA damage in yeast cells. Rad9 was originally found in budding yeast cells but a human homolog has also been found and studies have suggested that the molecular mechanisms of the S and G2 checkpoints are conserved in eukaryotes. Thus, what is found in yeast cells are likely to be similar in human cells.
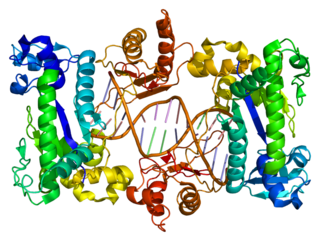
DNA polymerase iota is an enzyme that in humans is encoded by the POLI gene. It is found in higher eukaryotes, and is believed to have arisen from a gene duplication from Pol η. Pol ι, is a Y family polymerase that is involved in translesion synthesis. It can bypass 6-4 pyrimidine adducts and abasic sites and has a high frequency of wrong base incorporation. Like many other Y family polymerases Pol ι, has low processivity, a large DNA binding pocket and doesn't undergo conformational changes when DNA binds. These attributes are what allow Pol ι to carry out its task as a translesion polymerase. Pol ι only uses Hoogsteen base pairing, during DNA synthesis, it will add adenine opposite to thymine in the syn conformation and can add both cytosine and thymine in the anti conformation across guanine, which it flips to the syn conformation.
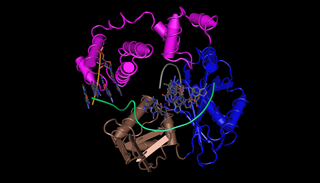
DNA polymerase lambda, also known as Pol λ, is an enzyme found in all eukaryotes. In humans, it is encoded by the POLL gene.

DNA topoisomerase 2-binding protein 1 (TOPBP1) is a scaffold protein that in humans is encoded by the TOPBP1 gene.

DNA polymerase eta, is a protein that in humans is encoded by the POLH gene.
Genome instability refers to a high frequency of mutations within the genome of a cellular lineage. These mutations can include changes in nucleic acid sequences, chromosomal rearrangements or aneuploidy. Genome instability does occur in bacteria. In multicellular organisms genome instability is central to carcinogenesis, and in humans it is also a factor in some neurodegenerative diseases such as amyotrophic lateral sclerosis or the neuromuscular disease myotonic dystrophy.

In molecular biology, kataegis describes a pattern of localized hypermutations identified in some cancer genomes, in which a large number of highly patterned basepair mutations occur in a small region of DNA. The mutational clusters are usually several hundred basepairs long, alternating between a long range of C→T substitutional pattern and a long range of G→A substitutional pattern. This suggests that kataegis is carried out on only one of the two template strands of DNA during replication. Compared to other cancer-related mutations, such as chromothripsis, kataegis is more commonly seen; it is not an accumulative process but likely happens during one cycle of replication.
DNA damage is an alteration in the chemical structure of DNA, such as a break in a strand of DNA, a nucleobase missing from the backbone of DNA, or a chemically changed base such as 8-OHdG. DNA damage can occur naturally or via environmental factors, but is distinctly different from mutation, although both are types of error in DNA. DNA damage is an abnormal chemical structure in DNA, while a mutation is a change in the sequence of base pairs. DNA damages cause changes in the structure of the genetic material and prevents the replication mechanism from functioning and performing properly. The DNA damage response (DDR) is a complex signal transduction pathway which recognizes when DNA is damaged and initiates the cellular response to the damage.
DNA polymerase IV is a prokaryotic polymerase that is involved in mutagenesis and is encoded by the dinB gene. It exhibits no 3′→5′ exonuclease (proofreading) activity and hence is error prone. In E. coli, DNA polymerase IV is involved in non-targeted mutagenesis. Pol IV is a Family Y polymerase expressed by the dinB gene that is switched on via SOS induction caused by stalled polymerases at the replication fork. During SOS induction, Pol IV production is increased tenfold and one of the functions during this time is to interfere with Pol III holoenzyme processivity. This creates a checkpoint, stops replication, and allows time to repair DNA lesions via the appropriate repair pathway. Another function of Pol IV is to perform translesion synthesis at the stalled replication fork like, for example, bypassing N2-deoxyguanine adducts at a faster rate than transversing undamaged DNA. Cells lacking dinB gene have a higher rate of mutagenesis caused by DNA damaging agents.

Sir Stephen Philip Jackson, FRS, FMedSci is the Frederick James Quick Professor of Biology. He is a senior group leader at the Cancer Research UK Cambridge Institute and associate group leader at the Gurdon Institute, University of Cambridge.

PrimPol is a protein encoded by the PRIMPOL gene in humans. PrimPol is a eukaryotic protein with both DNA polymerase and DNA Primase activities involved in translesion DNA synthesis. It is the first eukaryotic protein to be identified with priming activity using deoxyribonucleotides. It is also the first protein identified in the mitochondria to have translesion DNA synthesis activities.

Dame Sarah Catherine Gilbert FRS is an English vaccinologist who is a Professor of Vaccinology at the University of Oxford and co-founder of Vaccitech. She specialises in the development of vaccines against influenza and emerging viral pathogens. She led the development and testing of the universal flu vaccine, which underwent clinical trials in 2011.

The Jenner Institute is a research institute on the Old Road Campus in Headington, east Oxford, England. It was formed in November 2005 through a partnership between the University of Oxford and the UK Institute for Animal Health. It is associated with the Nuffield Department of Medicine, in the Medical Sciences Division of Oxford University. The institute receives charitable support from the Jenner Vaccine Foundation.

The Oxford–AstraZeneca COVID‑19 vaccine, sold under the brand names Covishield and Vaxzevria among others, is a viral vector vaccine for the prevention of COVID-19. It was developed in the United Kingdom by Oxford University and British-Swedish company AstraZeneca, using as a vector the modified chimpanzee adenovirus ChAdOx1. The vaccine is given by intramuscular injection. Studies carried out in 2020 showed that the efficacy of the vaccine is 76.0% at preventing symptomatic COVID-19 beginning at 22 days following the first dose and 81.3% after the second dose. A study in Scotland found that, for symptomatic COVID-19 infection after the second dose, the vaccine is 81% effective against the Alpha variant and 61% against the Delta variant.
References
- ↑ "Cell Division Cycle Laboratory". Imperial Cancer Research Fund. 2000. Archived from the original on 25 October 2000.
- 1 2 3 4 5 "Christ's College Magazine 2008". Issuu. Retrieved 24 April 2020.
- ↑ "Weekly Bulletin: Mayfield Grammar School" (PDF). Mayfield Grammar School . Retrieved 20 July 2021.
- ↑ "Churchill alumni listed in Queen's Birthday Honours". Churchill College, Cambridge . 11 June 2016. Retrieved 19 July 2021.
- ↑ Vialard, J E; Gilbert, C S; Green, C M; Lowndes, N F (1 October 1998). "The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage". The EMBO Journal. 17 (19): 5679–5688. doi:10.1093/emboj/17.19.5679. ISSN 0261-4189. PMC 1170896 . PMID 9755168.
- 1 2 3 "Dr Catherine Green PhD | Christs College Cambridge". www.christs.cam.ac.uk. Retrieved 24 April 2020.
- ↑ Oxford, N. D. M. (21 November 2014), Catherine Green: DNA replication and cancer , retrieved 24 April 2020
- ↑ "Department of Zoology, University of Cambridge, United Kingdom". Epigenesys. Retrieved 24 April 2020.
- 1 2 3 "Exeter Fellow Dr Catherine Green leads the production of a potential COVID-19 vaccine in Oxford". Exeter College. 6 April 2020. Retrieved 24 April 2020.
- ↑ "Catherine Green — Wellcome Centre for Human Genetics". www.well.ox.ac.uk. Retrieved 24 April 2020.
- ↑ "Hear from the Oxford COVID-19 vaccine team | Science Media Centre" . Retrieved 24 April 2020.
- 1 2 3 Hoare, Callum (2 April 2020). "Oxford University scientist tips miracle COVID-19 'neutraliser' for NHS frontline staff". Express.co.uk. Retrieved 24 April 2020.
- ↑ "Coronavirus vaccine: Professor behind trial tells James O'Brien what happens next". LBC. Retrieved 24 April 2020.
- ↑ "Covid-19: Oxford-AstraZeneca coronavirus vaccine approved for use in UK". BBC News. BBC. 30 December 2020. Retrieved 30 December 2020.
- ↑ "AstraZeneca withdraws Covid-19 vaccine, citing low demand". CNN . Retrieved 9 May 2024.
- ↑ Honigsbaum, Mark (11 July 2021). "Vaxxers by Sarah Gilbert and Catherine Green; Until Proven Safe by Geoff Manaugh and Nicola Twilley – reviews". The Guardian. Retrieved 12 June 2022.
- ↑ Ledford, Heidi (5 August 2021). "The COVID vaccine makers tell all". Nature. 596 (7870): 29–30. Bibcode:2021Natur.596...29L. doi:10.1038/d41586-021-02090-9. S2CID 236883504.
- ↑ "Catherine Green". Google Scholar . Retrieved 11 May 2024.
- ↑ Bienko, Marzena; Green, Catherine M.; Crosetto, Nicola; Rudolf, Fabian; Zapart, Grzegorz; Coull, Barry; Kannouche, Patricia; Wider, Gerhard; Peter, Matthias; Lehmann, Alan R.; Hofmann, Kay (16 December 2005). "Ubiquitin-Binding Domains in Y-Family Polymerases Regulate Translesion Synthesis". Science. 310 (5755): 1821–1824. Bibcode:2005Sci...310.1821B. doi:10.1126/science.1120615. ISSN 0036-8075. PMID 16357261. S2CID 10666348.
- ↑ Lehmann, Alan R.; Niimi, Atsuko; Ogi, Tomoo; Brown, Stephanie; Sabbioneda, Simone; Wing, Jonathan F.; Kannouche, Patricia L.; Green, Catherine M. (1 July 2007). "Translesion synthesis: Y-family polymerases and the polymerase switch". DNA Repair. Replication Fork Repair Processes. 6 (7): 891–899. doi:10.1016/j.dnarep.2007.02.003. ISSN 1568-7864. PMID 17363342.
- ↑ Gilbert, Christopher S; Green, Catherine M; Lowndes, Noel F (1 July 2001). "Budding Yeast Rad9 Is an ATP-Dependent Rad53 Activating Machine". Molecular Cell. 8 (1): 129–136. doi: 10.1016/S1097-2765(01)00267-2 . ISSN 1097-2765. PMID 11511366.