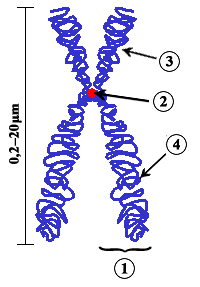
A chromosome is a long DNA molecule with part or all of the genetic material of an organism. Most eukaryotic chromosomes include packaging proteins called histones which, aided by chaperone proteins, bind to and condense the DNA molecule to maintain its integrity. These chromosomes display a complex three-dimensional structure, which plays a significant role in transcriptional regulation.

The centromere is the specialized DNA sequence of a chromosome that links a pair of sister chromatids. During mitosis, spindle fibers attach to the centromere via the kinetochore. Centromeres were first thought to be genetic loci that direct the behavior of chromosomes.

Meiosis is a special type of cell division of germ cells in sexually-reproducing organisms used to produce the gametes, such as sperm or egg cells. It involves two rounds of division that ultimately result in four cells with only one copy of each chromosome (haploid). Additionally, prior to the division, genetic material from the paternal and maternal copies of each chromosome is crossed over, creating new combinations of code on each chromosome. Later on, during fertilisation, the haploid cells produced by meiosis from a male and female will fuse to create a cell with two copies of each chromosome again, the zygote.

Cell division is the process by which a parent cell divides into two or more daughter cells. Cell division usually occurs as part of a larger cell cycle. In eukaryotes, there are two distinct types of cell division; a vegetative division, whereby each daughter cell is genetically identical to the parent cell (mitosis), and a reproductive cell division, whereby the number of chromosomes in the daughter cells is reduced by half to produce haploid gametes (meiosis). In cell biology, mitosis (/maɪˈtoʊsɪs/) is a part of the cell cycle, in which, replicated chromosomes are separated into two new nuclei. Cell division gives rise to genetically identical cells in which the total number of chromosomes is maintained. In general, mitosis is preceded by the S stage of interphase and is often followed by telophase and cytokinesis; which divides the cytoplasm, organelles and cell membrane of one cell into two new cells containing roughly equal shares of these cellular components. The different stages of Mitosis all together define the mitotic (M) phase of an animal cell cycle—the division of the mother cell into two genetically identical daughter cells. Meiosis results in four haploid daughter cells by undergoing one round of DNA replication followed by two divisions. Homologous chromosomes are separated in the first division, and sister chromatids are separated in the second division. Both of these cell division cycles are used in the process of sexual reproduction at some point in their life cycle. Both are believed to be present in the last eukaryotic common ancestor.
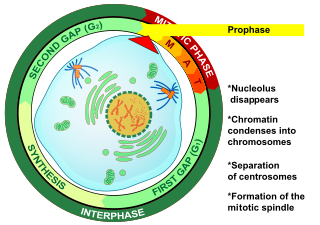
Prophase (from the Greek πρό, "before" and φάσις, "stage") is the first stage of cell division in both mitosis and meiosis. Beginning after interphase, DNA has already been replicated when the cell enters prophase. The main occurrences in prophase are the condensation of the chromatin reticulum and the disappearance of the nucleolus.
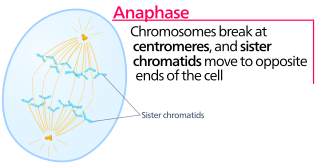
Anaphase, is the stage of mitosis after the process of metaphase, when replicated chromosomes are split and the newly-copied chromosomes are moved to opposite poles of the cell. Chromosomes also reach their overall maximum condensation in late anaphase, to help chromosome segregation and the re-formation of the nucleus.

In cell biology, the spindle apparatus refers to the cytoskeletal structure of eukaryotic cells that forms during cell division to separate sister chromatids between daughter cells. It is referred to as the mitotic spindle during mitosis, a process that produces genetically identical daughter cells, or the meiotic spindle during meiosis, a process that produces gametes with half the number of chromosomes of the parent cell.

A couple of homologous chromosomes, or homologs, are a set of one maternal and one paternal chromosome that pair up with each other inside a cell during fertilization. Homologs have the same genes in the same loci where they provide points along each chromosome which enable a pair of chromosomes to align correctly with each other before separating during meiosis. This is the basis for Mendelian inheritance which characterizes inheritance patterns of genetic material from an organism to its offspring parent developmental cell at the given time and area.

The spindle checkpoint, also known as the metaphase-to-anaphase transition, the spindle assembly checkpoint (SAC), the metaphase checkpoint, or the mitotic checkpoint, is a cell cycle checkpoint during mitosis or meiosis that prevents the separation of the duplicated chromosomes (anaphase) until each chromosome is properly attached to the spindle. To achieve proper segregation, the two kinetochores on the sister chromatids must be attached to opposite spindle poles. Only this pattern of attachment will ensure that each daughter cell receives one copy of the chromosome. The defining biochemical feature of this checkpoint is the stimulation of the anaphase-promoting complex by M-phase cyclin-CDK complexes, which in turn causes the proteolytic destruction of cyclins and proteins that hold the sister chromatids together.

A kinetochore is a disc-shaped protein structure associated with duplicated chromatids in eukaryotic cells where the spindle fibers attach during cell division to pull sister chromatids apart. The kinetochore assembles on the centromere and links the chromosome to microtubule polymers from the mitotic spindle during mitosis and meiosis. The term kinetochore was first used in a footnote in a 1934 Cytology book by Lester W. Sharp and commonly accepted in 1936. Sharp's footnote reads: "The convenient term kinetochore has been suggested to the author by J. A. Moore", likely referring to John Alexander Moore who had joined Columbia University as a freshman in 1932.

An isochromosome is an unbalanced structural abnormality in which the arms of the chromosome are mirror images of each other. The chromosome consists of two copies of either the long (q) arm or the short (p) arm because isochromosome formation is equivalent to a simultaneous duplication and deletion of genetic material. Consequently, there is partial trisomy of the genes present in the isochromosome and partial monosomy of the genes in the lost arm.
SMC complexes represent a large family of ATPases that participate in many aspects of higher-order chromosome organization and dynamics. SMC stands for Structural Maintenance of Chromosomes.
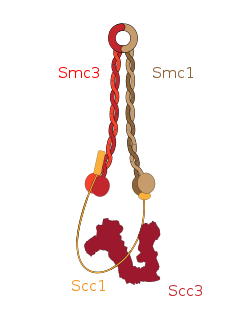
Cohesin is a protein complex that mediates sister chromatid cohesion, homologous recombination and DNA looping. Cohesin is formed of SMC3, SMC1, SCC1 and SCC3. Cohesin holds sister chromatids together after DNA replication until anaphase when removal of cohesin leads to separation of sister chromatids. The complex forms a ring-like structure and it is believed that sister chromatids are held together by entrapment inside the cohesin ring. Cohesin is a member of the SMC family of protein complexes which includes Condensin, MukBEF and SMC-ScpAB.
This glossary of genetics is a list of definitions of terms and concepts commonly used in the study of genetics and related disciplines in biology, including molecular biology and evolutionary biology. It is intended as introductory material for novices; for more specific and technical detail, see the article corresponding to each term. For related terms, see Glossary of evolutionary biology.

Aurora B kinase is a protein that functions in the attachment of the mitotic spindle to the centromere.

Centromere protein A, also known as CENPA, is a protein which in humans is encoded by the CENPA gene. CENPA is a histone H3 variant which is the critical factor determining the kinetochore position(s) on each chromosome in most eukaryotes including humans.

Centromere protein C 1 is a protein that in humans is encoded by the CENPC1 gene.
Quantitative Fluorescent in situ hybridization (Q-FISH) is a cytogenetic technique based on the traditional FISH methodology. In Q-FISH, the technique uses labelled synthetic DNA mimics called peptide nucleic acid (PNA) oligonucleotides to quantify target sequences in chromosomal DNA using fluorescent microscopy and analysis software. Q-FISH is most commonly used to study telomere length, which in vertebrates are repetitive hexameric sequences (TTAGGG) located at the distal end of chromosomes. Telomeres are necessary at chromosome ends to prevent DNA-damage responses as well as genome instability. To this day, the Q-FISH method continues to be utilized in the field of telomere research.

Neocentromeres are new centromeres that form at a place on the chromosome that is usually not centromeric. They typically arise due to disruption of the normal centromere. These neocentromeres should not be confused with “knobs”, which were also described as “neocentromeres” in maize in the 1950s. Unlike most normal centromeres, neocentromeres do not contain satellite sequences that are highly repetitive but instead consist of unique sequences. Despite this, most neocentromeres are still able to carry out the functions of normal centromeres in regulating chromosome segregation and inheritance. This raises many questions on what is necessary versus what is sufficient for constituting a centromere.
Holocentric chromosomes are chromosomes that possess multiple kinetochores along their length rather than the single centromere typical of other chromosomes. They were first described in cytogenetic experiments in 1935. Since this first observation, the term holocentric chromosome has referred to chromosomes that: i) lack the primary constriction corresponding to the centromere observed in monocentric chromosomes; and ii) possess multiple kinetochores dispersed along the entire chromosomal axis, such that microtubules bind to the chromosome along its entire length and move broadside to the pole from the metaphase plate. Holocentric chromosomes are also termed holokinetic, because, during cell division, the sister chromatids move apart in parallel and do not form the classical V-shaped figures typical of monocentric chromosomes.