
Cytochromes are redox-active proteins containing a heme, with a central Fe atom at its core, as a cofactor. They are involved in electron transport chain and redox catalysis. They are classified according to the type of heme and its mode of binding. Four varieties are recognized by the International Union of Biochemistry and Molecular Biology (IUBMB), cytochromes a, cytochromes b, cytochromes c and cytochrome d. Cytochrome function is linked to the reversible redox change from ferrous to the ferric oxidation state of the iron found in the heme core. In addition to the classification by the IUBMB into four cytochrome classes, several additional classifications such as cytochrome o and cytochrome P450 can be found in biochemical literature.

Oxidative phosphorylation is the metabolic pathway in which cells use enzymes to oxidize nutrients, thereby releasing the chemical energy stored within in order to produce adenosine triphosphate (ATP). In most eukaryotes, this takes place inside mitochondria. Almost all aerobic organisms carry out oxidative phosphorylation. This pathway is so pervasive because the energy of the double bond of oxygen is so much higher than the energy of the double bond in carbon dioxide or in pairs of single bonds in organic molecules observed in alternative fermentation processes such as anaerobic glycolysis.

Peroxidases or peroxide reductases are a large group of enzymes which play a role in various biological processes. They are named after the fact that they commonly break up peroxides.

The enzyme cytochrome c oxidase or Complex IV, EC 1.9.3.1, is a large transmembrane protein complex found in bacteria, archaea, and the mitochondria of eukaryotes.
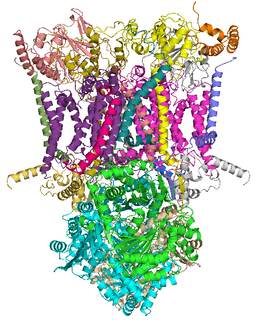
The coenzyme Q : cytochrome c – oxidoreductase, sometimes called the cytochrome bc1 complex, and at other times complex III, is the third complex in the electron transport chain, playing a critical role in biochemical generation of ATP. Complex III is a multisubunit transmembrane protein encoded by both the mitochondrial and the nuclear genomes. Complex III is present in the mitochondria of all animals and all aerobic eukaryotes and the inner membranes of most eubacteria. Mutations in Complex III cause exercise intolerance as well as multisystem disorders. The bc1 complex contains 11 subunits, 3 respiratory subunits, 2 core proteins and 6 low-molecular weight proteins.

Heme or haem is a substance precursive to hemoglobin, which is necessary to bind oxygen in the bloodstream. Haem is biosynthesized in both the bone marrow and the liver.

Cytochrome c peroxidase, or CCP, is a water-soluble heme-containing enzyme of the peroxidase family that takes reducing equivalents from cytochrome c and reduces hydrogen peroxide to water:
Ferredoxins are iron–sulfur proteins that mediate electron transfer in a range of metabolic reactions. The term "ferredoxin" was coined by D.C. Wharton of the DuPont Co. and applied to the "iron protein" first purified in 1962 by Mortenson, Valentine, and Carnahan from the anaerobic bacterium Clostridium pasteurianum.

In biochemistry, flavin adenine dinucleotide (FAD) is a redox-active coenzyme associated with various proteins, which is involved with several enzymatic reactions in metabolism. A flavoprotein is a protein that contains a flavin group, which may be in the form of FAD or flavin mononucleotide (FMN). Many flavoproteins are known: components of the succinate dehydrogenase complex, α-ketoglutarate dehydrogenase, and a component of the pyruvate dehydrogenase complex.

The cytochrome b6f complex is an enzyme found in the thylakoid membrane in chloroplasts of plants, cyanobacteria, and green algae, that catalyzes the transfer of electrons from plastoquinol to plastocyanin. The reaction is analogous to the reaction catalyzed by cytochrome bc1 of the mitochondrial electron transport chain. During photosynthesis, the cytochrome b6f complex is one step along the chain that transfers electrons from Photosystem II to Photosystem I, and at the same time pumps protons into the thylakoid space that contribute to create an electrochemical (energy) gradient which is later used to synthesize ATP from ADP.

The Q cycle describes a series of reactions that describe how the sequential oxidation and reduction of the lipophilic electron carrier, Coenzyme Q10 (CoQ10), between the ubiquinol and ubiquinone forms, can result in the net movement of protons across a lipid bilayer.
Nitrite reductase refers to any of several classes of enzymes that catalyze the reduction of nitrite. There are two classes of NIR's. A multi haem enzyme reduces NO2− to a variety of products. Copper containing enzymes carry out a single electron transfer to produce nitric oxide.
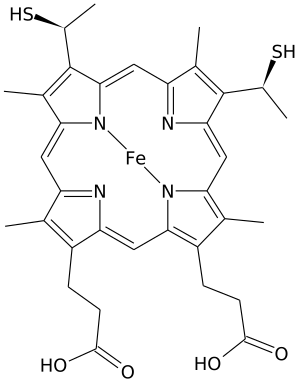
Heme C is an important kind of heme.
In enzymology, a NADH peroxidase (EC 1.11.1.1) is an enzyme that catalyzes the chemical reaction

Cytochromes c cytochromes, or heme-containing proteins, that have heme C covalently attached to the peptide backbone via one or two thioether bonds. These bonds are in most cases part of a specific Cys-X-X-Cys-His (CXXCH) binding motif, where X denotes a miscellaneous amino acid. Two thioether bonds of cysteine residues bind to the vinyl sidechains of heme, and the histidine residue coordinates one axial binding site of the heme iron. Less common binding motifs can include a single thioether linkage, a lysine or a methionine instead of the axial histidine or a CXnCH binding motif with n>2. The second axial site of the iron can be coordinated by amino acids of the protein, substrate molecules or water. Cytochromes c possess a wide range of properties and function as electron transfer proteins or catalyse chemical reactions involving redox processes. A prominent member of this family is mitochondrial cytochrome c.

Dual oxidase 1, also known as DUOX1 or ThOX1, is an enzyme which in humans is encoded by the DUOX1 gene. DUOX1 was first identified in the mammalian thyroid gland. In humans, two isoforms are found; hDUOX1 and hDUOX2. Human DUOX protein localization is not exclusive to thyroid tissue; hDUOX1 is prominent in airway epithelial cells and hDUOX2 in the salivary glands and gastrointestinal tract.

Azurin is a small, periplasmic, bacterial blue copper protein found in Pseudomonas, Bordetella, or Alcaligenes bacteria. Azurin moderates single-electron transfer between enzymes associated with the cytochrome chain by undergoing oxidation-reduction between Cu(I) and Cu(II). Each monomer of an azurin tetramer has a molecular weight of approximately 14kDa, contains a single copper atom, is intensively blue, and has a fluorescence emission band centered at 308 nm.
Haem peroxidases (or heme peroxidases) are haem-containing enzymes that use hydrogen peroxide as the electron acceptor to catalyse a number of oxidative reactions. Most haem peroxidases follow the reaction scheme:

Flavocytochrome c sulfide dehydrogenase, also known as Sulfide-cytochrome-c reductase (flavocytochrome c) (EC 1.8.2.3), is an enzyme with systematic name hydrogen-sulfide:flavocytochrome c oxidoreductase. It is found in sulfur-oxidising bacteria such as the purple phototrophic bacteria Allochromatium vinosum. This enzyme catalyses the following chemical reaction:

Eosinophil peroxidase is an enzyme found within the eosinophil granulocytes, innate immune cells of humans and mammals. This oxidoreductase protein is encoded by the gene EPX, expressed within these myeloid cells. EPO shares many similarities with its orthologous peroxidases, myeloperoxidase (MPO), lactoperoxidase (LPO), and thyroid peroxidase (TPO). The protein is concentrated in secretory granules within eosinophils. Eosinophil peroxidase is a heme peroxidase, its activities including the oxidation of halide ions to bacteriocidal reactive oxygen species, the cationic disruption of bacterial cell walls, and the post-translational modification of protein amino acid residues.

















