
A chromosome is a long DNA molecule with part or all of the genetic material of an organism. Most eukaryotic chromosomes include packaging proteins called histones which, aided by chaperone proteins, bind to and condense the DNA molecule to maintain its integrity. These chromosomes display a complex three-dimensional structure, which plays a significant role in transcriptional regulation.
Chromatin is a complex of DNA and protein found in eukaryotic cells. The primary function is to package long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important roles in reinforcing the DNA during cell division, preventing DNA damage, and regulating gene expression and DNA replication. During mitosis and meiosis, chromatin facilitates proper segregation of the chromosomes in anaphase; the characteristic shapes of chromosomes visible during this stage are the result of DNA being coiled into highly condensed chromatin.

Deoxyribonucleic acid is a polymer composed of two polynucleotide chains that coil around each other to form a double helix carrying genetic instructions for the development, functioning, growth and reproduction of all known organisms and many viruses. DNA and ribonucleic acid (RNA) are nucleic acids. Alongside proteins, lipids and complex carbohydrates (polysaccharides), nucleic acids are one of the four major types of macromolecules that are essential for all known forms of life.

In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn are wrapped into 30-nanometer fibers that form tightly packed chromatin. Histones prevent DNA from becoming tangled and protect it from DNA damage. In addition, histones play important roles in gene regulation and DNA replication. Without histones, unwound DNA in chromosomes would be very long. For example, each human cell has about 1.8 meters of DNA if completely stretched out; however, when wound about histones, this length is reduced to about 90 micrometers (0.09 um) of 30 nm diameter chromatin fibers.
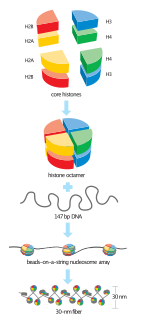
A nucleosome is the basic structural unit of DNA packaging in eukaryotes. The structure of a nucleosome consists of a segment of DNA wound around eight histone proteins and resembles thread wrapped around a spool. The nucleosome is the fundamental subunit of chromatin. Each nucleosome is composed of a little less than two turns of DNA wrapped around a set of eight proteins called histones, which are known as a histone octamer. Each histone octamer is composed of two copies each of the histone proteins H2A, H2B, H3, and H4.
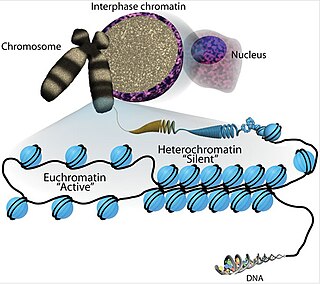
Euchromatin is a lightly packed form of chromatin that is enriched in genes, and is often under active transcription. Euchromatin stands in contrast to heterochromatin, which is tightly packed and less accessible for transcription. 92% of the human genome is euchromatic.

A histone octamer is the eight protein complex found at the center of a nucleosome core particle. It consists of two copies of each of the four core histone proteins. The octamer assembles when a tetramer, containing two copies of both H3 and H4, complexes with two H2A/H2B dimers. Each histone has both an N-terminal tail and a C-terminal histone-fold. Both of these key components interact with DNA in their own way through a series of weak interactions, including hydrogen bonds and salt bridges. These interactions keep the DNA and histone octamer loosely associated and ultimately allow the two to re-position or separate entirely.
A histone fold is a structurally conserved motif found near the C-terminus in every core histone sequence in a histone octamer responsible for the binding of histones into heterodimers.

DNA-binding proteins are proteins that have DNA-binding domains and thus have a specific or general affinity for single- or double-stranded DNA. Sequence-specific DNA-binding proteins generally interact with the major groove of B-DNA, because it exposes more functional groups that identify a base pair. However, there are some known minor groove DNA-binding ligands such as netropsin, distamycin, Hoechst 33258, pentamidine, DAPI and others.
Histone H1 is one of the five main histone protein families which are components of chromatin in eukaryotic cells. Though highly conserved, it is nevertheless the most variable histone in sequence across species.

Histone H2A is one of the five main histone proteins involved in the structure of chromatin in eukaryotic cells.
Histone H2B is one of the 5 main histone proteins involved in the structure of chromatin in eukaryotic cells. Featuring a main globular domain and long N-terminal and C-terminal tails, H2B is involved with the structure of the nucleosomes.

Eukaryotic DNA replication is a conserved mechanism that restricts DNA replication to once per cell cycle. Eukaryotic DNA replication of chromosomal DNA is central for the duplication of a cell and is necessary for the maintenance of the eukaryotic genome.

The solenoid structure of chromatin is a model for the structure of the 30 nm fibre. It is a secondary chromatin structure which helps to package eukaryotic DNA into the nucleus.

Histone H3.1 is a protein that in humans is encoded by the HIST1H3B gene.

Histone H3.2 is a protein that in humans is encoded by the HIST2H3C gene.

Histone H2A-Bbd type 2/3 also known as H2A Barr body-deficient is a histone protein that in humans is encoded by the H2AFB2 gene.

DNA condensation refers to the process of compacting DNA molecules in vitro or in vivo. Mechanistic details of DNA packing are essential for its functioning in the process of gene regulation in living systems. Condensed DNA often has surprising properties, which one would not predict from classical concepts of dilute solutions. Therefore, DNA condensation in vitro serves as a model system for many processes of physics, biochemistry and biology. In addition, DNA condensation has many potential applications in medicine and biotechnology.

Nucleic acidquaternary structure refers to the interactions between separate nucleic acid molecules, or between nucleic acid molecules and proteins. The concept is analogous to protein quaternary structure, but as the analogy is not perfect, the term is used to refer to a number of different concepts in nucleic acids and is less commonly encountered. Similarly other biomolecules such as proteins, nucleic acids have four levels of structural arrangement: primary, secondary, tertiary, and quaternary structure. Primary structure is the linear sequence of nucleotides, secondary structure involves small local folding motifs, and tertiary structure is the 3D folded shape of nucleic acid molecule. In general, quaternary structure refers to 3D interactions between multiple subunits. In the case of nucleic acids, quaternary structure refers to interactions between multiple nucleic acid molecules or between nucleic acids and proteins. Nucleic acid quaternary structure is important for understanding DNA, RNA, and gene expression because quaternary structure can impact function. For example, when DNA is packed into chromatin, therefore exhibiting a type of quaternary structure, gene transcription will be inhibited.
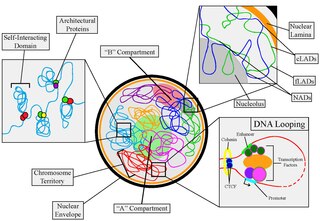
Nuclear organization refers to the spatial distribution of chromatin within a cell nucleus. There are many different levels and scales of nuclear organisation. Chromatin is a higher order structure of DNA.

















