Related Research Articles

Influenza A virus (IAV) is a pathogen that causes the flu in birds and some mammals, including humans. It is an RNA virus whose subtypes have been isolated from wild birds. Occasionally, it is transmitted from wild to domestic birds, and this may cause severe disease, outbreaks, or human influenza pandemics.

Influenza hemagglutinin (HA) or haemagglutinin[p] is a homotrimeric glycoprotein found on the surface of influenza viruses and is integral to its infectivity.

Orthomyxoviridae is a family of negative-sense RNA viruses. It includes seven genera: Alphainfluenzavirus, Betainfluenzavirus, Gammainfluenzavirus, Deltainfluenzavirus, Isavirus, Thogotovirus, and Quaranjavirus. The first four genera contain viruses that cause influenza in birds and mammals, including humans. Isaviruses infect salmon; the thogotoviruses are arboviruses, infecting vertebrates and invertebrates. The Quaranjaviruses are also arboviruses, infecting vertebrates (birds) and invertebrates (arthropods).
Antigenic drift is a kind of genetic variation in viruses, arising from the accumulation of mutations in the virus genes that code for virus-surface proteins that host antibodies recognize. This results in a new strain of virus particles that is not effectively inhibited by the antibodies that prevented infection by previous strains. This makes it easier for the changed virus to spread throughout a partially immune population. Antigenic drift occurs in both influenza A and influenza B viruses.

Influenza vaccines, colloquially known as flu shots, are vaccines that protect against infection by influenza viruses. New versions of the vaccines are developed twice a year, as the influenza virus rapidly changes. While their effectiveness varies from year to year, most provide modest to high protection against influenza. Vaccination against influenza began in the 1930s, with large-scale availability in the United States beginning in 1945.

Swine influenza is an infection caused by any of several types of swine influenza viruses. Swine influenza virus (SIV) or swine-origin influenza virus (S-OIV) refers to any strain of the influenza family of viruses that is endemic in pigs. As of 2009, identified SIV strains include influenza C and the subtypes of influenza A known as H1N1, H1N2, H2N1, H3N1, H3N2, and H2N3.

Original antigenic sin, also known as antigenic imprinting, the Hoskins effect, immunological imprinting, or primary addiction is the propensity of the immune system to preferentially use immunological memory based on a previous infection when a second slightly different version of that foreign pathogen is encountered. This leaves the immune system "trapped" by the first response it has made to each antigen, and unable to mount potentially more effective responses during subsequent infections. Antibodies or T-cells induced during infections with the first variant of the pathogen are subject to repertoire freeze, a form of original antigenic sin.
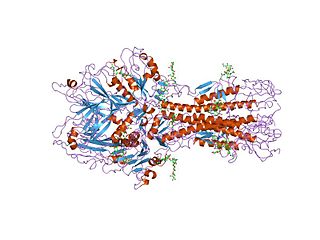
Hemagglutinin esterase (HEs) is a glycoprotein that certain enveloped viruses possess and use as an invading mechanism. HEs helps in the attachment and destruction of certain sialic acid receptors that are found on the host cell surface. Viruses that possess HEs include influenza C virus, toroviruses, and coronaviruses of the subgenus Embecovirus. HEs is a dimer transmembrane protein consisting of two monomers, each monomer is made of three domains. The three domains are: membrane fusion, esterase, and receptor binding domains.

H5N1 genetic structure is the molecular structure of the H5N1 virus's RNA.

H5N1 clinical trials are clinical trials concerning H5N1 vaccines, which are intended to provide immunization to influenza A virus subtype H5N1. They are intended to discover pharmacological effects and identify any adverse reactions the vaccines may achieve in humans.
Antigenic variation or antigenic alteration refers to the mechanism by which an infectious agent such as a protozoan, bacterium or virus alters the proteins or carbohydrates on its surface and thus avoids a host immune response, making it one of the mechanisms of antigenic escape. It is related to phase variation. Antigenic variation not only enables the pathogen to avoid the immune response in its current host, but also allows re-infection of previously infected hosts. Immunity to re-infection is based on recognition of the antigens carried by the pathogen, which are "remembered" by the acquired immune response. If the pathogen's dominant antigen can be altered, the pathogen can then evade the host's acquired immune system. Antigenic variation can occur by altering a variety of surface molecules including proteins and carbohydrates. Antigenic variation can result from gene conversion, site-specific DNA inversions, hypermutation, or recombination of sequence cassettes. The result is that even a clonal population of pathogens expresses a heterogeneous phenotype. Many of the proteins known to show antigenic or phase variation are related to virulence.

Human mortality from H5N1 or the human fatality ratio from H5N1 or the case-fatality rate of H5N1 is the ratio of the number of confirmed human deaths resulting from confirmed cases of transmission and infection of H5N1 to the number of those confirmed cases. For example, if there are 100 confirmed cases of humans infected with H5N1 and 10 die, then there is a 10% human fatality ratio. H5N1 flu is a concern due to the global spread of H5N1 that constitutes a pandemic threat. The majority of H5N1 flu cases have been reported in southeast and east Asia. The case-fatality rate is central to pandemic planning. Estimates of case-fatality (CF) rates for past influenza pandemics have ranged from to 2-3% for the 1918 pandemic to about 0.6% for the 1957 pandemic to 0.2% for the 1968 pandemic. As of 2008, the official World Health Organization estimate for the case-fatality rate for the outbreak of H5N1 avian influenza was approximately 60%. Public health officials in Ontario, Canada argue that the true case-fatality rate could be lower, pointing to studies suggesting it could be 14-33%, and warned that it was unlikely to be as low as the 0.1–0.4% rate that was built into many pandemic plans.

Influenza, commonly known as "the flu" or just "flu", is an infectious disease caused by influenza viruses. Symptoms range from mild to severe and often include fever, runny nose, sore throat, muscle pain, headache, coughing, and fatigue. These symptoms begin from one to four days after exposure to the virus and last for about 2–8 days. Diarrhea and vomiting can occur, particularly in children. Influenza may progress to pneumonia, which can be caused by the virus or by a subsequent bacterial infection. Other complications of infection include acute respiratory distress syndrome, meningitis, encephalitis, and worsening of pre-existing health problems such as asthma and cardiovascular disease.
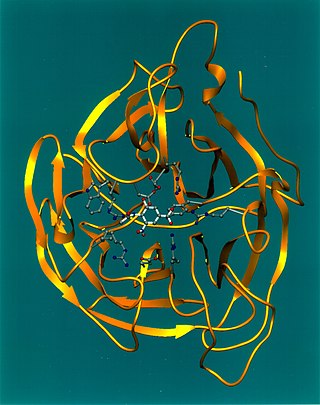
Viral neuraminidase is a type of neuraminidase found on the surface of influenza viruses that enables the virus to be released from the host cell. Neuraminidases are enzymes that cleave sialic acid groups from glycoproteins. Viral neuraminidase was discovered by Alfred Gottschalk at the Walter and Eliza Hall Institute in 1957. Neuraminidase inhibitors are antiviral agents that inhibit influenza viral neuraminidase activity and are of major importance in the control of influenza.
CR6261 is a monoclonal antibody that binds to a broad range of the influenza virus including the 1918 "Spanish flu" (SC1918/H1) and to a virus of the H5N1 class of avian influenza that jumped from chickens to a human in Vietnam in 2004 (Viet04/H5). In contrast to most antibodies generated by exposure to influenza, which can only neutralize a few strains from within a single virus subtype, CR6261 neutralizes numerous strains from multiple subtypes. CR6261 recognizes a highly conserved helical region in the membrane-proximal stem of hemagglutinin, the predominant protein on the surface of the influenza virus. Based upon the conservation of the amino acid sequence on this part of hemagglutinin, CR6261 is predicted to neutralize roughly 50% of all flu viruses. It was found by The Scripps Research Institute and the Dutch biopharmaceutical company, Crucell.

The pandemic H1N1/09 virus is a swine origin influenza A virus subtype H1N1 strain that was responsible for the 2009 swine flu pandemic. This strain is often called swine flu by the public media. For other names, see the Nomenclature section below.
A neutralizing antibody (NAb) is an antibody that defends a cell from a pathogen or infectious particle by neutralizing any effect it has biologically. Neutralization renders the particle no longer infectious or pathogenic. Neutralizing antibodies are part of the humoral response of the adaptive immune system against viruses, intracellular bacteria and microbial toxin. By binding specifically to surface structures (antigen) on an infectious particle, neutralizing antibodies prevent the particle from interacting with its host cells it might infect and destroy.

A H5N1 vaccine is an influenza vaccine intended to provide immunization to influenza A virus subtype H5N1.
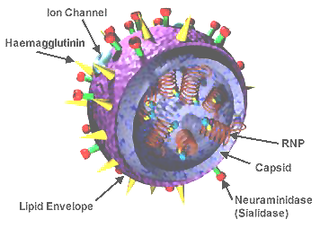
A universal flu vaccine is a flu vaccine that is effective against all influenza strains regardless of the virus sub type, antigenic drift or antigenic shift. Hence it should not require modification from year to year. As of 2021 no universal flu vaccine had been approved for general use, several were in development, and one was in clinical trial.
Type A influenza vaccine is for the prevention of infection of influenza A virus and also the influenza-related complications. Different monovalent type A influenza vaccines have been developed for different subtypes of influenza A virus including H1N1 and H5N1. Both intramuscular injection or intranasal spray are available on market. Unlike the seasonal influenza vaccines which are used annually, they are usually used during the outbreak of certain strand of subtypes of influenza A. Common adverse effects includes injection site reaction and local tenderness. Incidences of headache and myalgia were also reported with H1N1 whereas cases of fever has also been demonstrated with H5N1 vaccines. It is stated that immunosuppressant therapies would reduce the therapeutic effects of vaccines and that people with egg allergy should go for the egg-free preparations.
References
- ↑ Gallagher, James (29 July 2011). "'Super antibody' fights off flu". BBC.
- ↑ Laurance, Jeremy (29 July 2011). "Scientists hail the prospect of a universal vaccine for flu". Independent.
- ↑ Chan AL (28 July 2011). "Universal Flu Vaccine On The Horizon: Researchers Find 'Super Antibody'". Huff Post.
- 1 2 Corti, D; Voss, J; Gamblin, SJ; Codoni, G; Macagno, A; Jarrossay, D; Vachieri, SG; Pinna, D; Minola, A; Vanzetta, F; Silacci, C; Fernandez-Rodriguez, BM; Agatic, G; Bianchi, S; Giacchetto-Sasselli, I; Calder, L; Sallusto, F; Collins, P; Haire, LF; Temperton, N; Langedijk, JP; Skehel, JJ; Lanzavecchia, A (12 August 2011). "A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins". Science Magazine. 333 (6044): 850–6. Bibcode:2011Sci...333..850C. doi:10.1126/science.1205669. PMID 21798894. S2CID 5086468.
- ↑ Kelland, Kate (28 July 2011). "Flu 'super antibody' may bring universal shot closer". Reuters.
- 1 2 Russell, CJ (20 October 2011). "Stalking influenza diversity with a universal antibody" (PDF). The New England Journal of Medicine. 365 (16): 1541–2. doi:10.1056/NEJMcibr1109447. PMID 22010922.