In human genetics, the Y-chromosomal most recent common ancestor is the patrilineal most recent common ancestor (MRCA) from whom all currently living humans are descended. He is the most recent male from whom all living humans are descended through an unbroken line of their male ancestors. The term Y-MRCA reflects the fact that the Y chromosomes of all currently living human males are directly derived from the Y chromosome of this remote ancestor. The analogous concept of the matrilineal most recent common ancestor is known as "Mitochondrial Eve", the most recent woman from whom all living humans are descended matrilineally. As with "Mitochondrial Eve", the title of "Y-chromosomal Adam" is not permanently fixed to a single individual, but can advance over the course of human history as paternal lineages become extinct.
Genetics and archaeogenetics of South Asia is the study of the genetics and archaeogenetics of the ethnic groups of South Asia. It aims at uncovering these groups' genetic histories. The geographic position of the Indian subcontinent makes its biodiversity important for the study of the early dispersal of anatomically modern humans across Asia.
Haplogroup I (M170) is a Y-chromosome DNA haplogroup. It is a subgroup of haplogroup IJ, which itself is a derivative of the haplogroup IJK. Subclades I1 and I2 can be found in most present-day European populations, with peaks in some Northern European and Southeastern European countries.

Haplogroup J-M304, also known as J, is a human Y-chromosome DNA haplogroup. It is believed to have evolved in Western Asia. The clade spread from there during the Neolithic, primarily into North Africa, the Horn of Africa, the Socotra Archipelago, the Caucasus, Europe, Anatolia, Central Asia, South Asia, and Southeast Asia.

E-M215, also known as E1b1b-M215, is a major human Y-chromosome DNA haplogroup. E-M215 has two basal branches, E-M35 and E-M281. E-M35 is primarily distributed in North Africa and the Horn of Africa, and occurs at moderate frequencies in the Middle East, Europe, and Southern Africa. E-M281 occurs at a low frequency in Ethiopia.
Haplogroup E-M96 is a human Y-chromosome DNA haplogroup. It is one of the two main branches of the older and ancestral haplogroup DE, the other main branch being haplogroup D. The E-M96 clade is divided into two main subclades: the more common E-P147, and the less common E-M75.
Haplogroup K or K-M9 is a genetic lineage within human Y-chromosome DNA haplogroup. A sublineage of haplogroup IJK, K-M9, and its descendant clades represent a geographically widespread and diverse haplogroup. The lineages have long been found among males on every continent except Antarctica.
Haplogroup P also known as P-F5850 or K2b2 is a Y-chromosome DNA haplogroup in human genetics. P-F5850 is a branch of K2b, which is a branch of Haplogroup K2 (K-M526).
Haplogroup R, or R-M207, is a Y-chromosome DNA haplogroup. It is both numerous and widespread amongst modern populations.
In human genetics, Haplogroup O-M268, also known as O1b, is a Y-chromosome DNA haplogroup. Haplogroup O-M268 is a primary subclade of haplogroup O-F265, itself a primary descendant branch of Haplogroup O-M175.
In human genetics, Haplogroup O-M119 is a Y-chromosome DNA haplogroup. Haplogroup O-M119 is a descendant branch of haplogroup O-F265 also known as O1a, one of two extant primary subclades of Haplogroup O-M175. The same clade previously has been labeled as O-MSY2.2.
Haplogroup DE is a human Y-chromosome DNA haplogroup. It is defined by the single nucleotide polymorphism (SNP) mutations, or UEPs, M1(YAP), M145(P205), M203, P144, P153, P165, P167, P183. DE is unique because it is distributed in several geographically distinct clusters. An immediate subclade, haplogroup D, is mainly found in East Asia, parts of Central Asia, and the Andaman Islands, but also sporadically in West Africa and West Asia. The other immediate subclade, haplogroup E, is common in Africa, and to a lesser extent the Middle East and southern Europe.
The various ethnolinguistic groups found in the Caucasus, Central Asia, Europe, the Middle East, North Africa and/or South Asia demonstrate differing rates of particular Y-DNA haplogroups.
Haplogroup E-V68, also known as E1b1b1a, is a major human Y-chromosome DNA haplogroup found in North Africa, the Horn of Africa, Western Asia and Europe. It is a subclade of the larger and older haplogroup, known as E1b1b or E-M215. The E1b1b1a lineage is identified by the presence of a single nucleotide polymorphism (SNP) mutation on the Y chromosome, which is known as V68. It is a subject of discussion and study in genetics as well as genetic genealogy, archaeology, and historical linguistics.

The peopling of India refers to the migration of Homo sapiens into the Indian subcontinent. Anatomically modern humans settled India in multiple waves of early migrations, over tens of millennia. The first migrants came with the Coastal Migration/Southern Dispersal 65,000 years ago, whereafter complex migrations within South and Southeast Asia took place. West-Asian (Iranian) hunter-gatherers migrated to South Asia after the Last Glacial Period but before the onset of farming. Together with ancient South Asian hunter-gatherers they formed the population of the Indus Valley Civilisation (IVC).
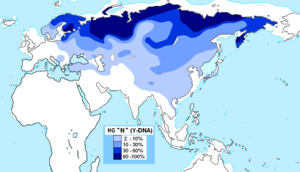
Y-DNA haplogroups in populations of Europe are haplogroups of the male Y-chromosome found in European populations.

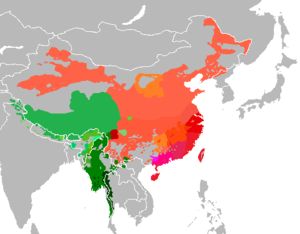
The tables below provide statistics on the human Y-chromosome DNA haplogroups most commonly found among ethnolinguistic groups and populations from East and South-East Asia.

The Munda peoples of eastern and central parts of the Indian subcontinent are any of several Munda speaking ethno-linguistic groups of Austro-asiatic language family, formerly also known as Kolarian, and spoken by about nine million people.
Various genetic studies on Filipinos have been performed, to analyze the population genetics of the various ethnic groups in the Philippines.
This article summarizes the genetic makeup and population history of East Asian peoples and their connection to genetically related populations, as well as Oceanians and partly, Central Asians and South Asians, which are collectively referred to as "East Eurasians" in population genomics.