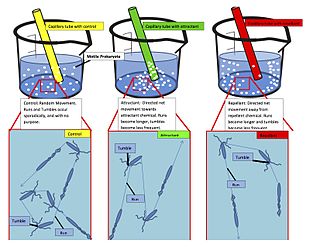
Chemotaxis is the movement of an organism or entity in response to a chemical stimulus. Somatic cells, bacteria, and other single-cell or multicellular organisms direct their movements according to certain chemicals in their environment. This is important for bacteria to find food by swimming toward the highest concentration of food molecules, or to flee from poisons. In multicellular organisms, chemotaxis is critical to early development and development as well as in normal function and health. In addition, it has been recognized that mechanisms that allow chemotaxis in animals can be subverted during cancer metastasis. The aberrant chemotaxis of leukocytes and lymphocytes also contribute to inflammatory diseases such as atherosclerosis, asthma, and arthritis. Sub-cellular components, such as the polarity patch generated by mating yeast, may also display chemotactic behavior.

Arabidopsis thaliana, the thale cress, mouse-ear cress or arabidopsis, is a small plant from the mustard family (Brassicaceae), native to Eurasia and Africa. Commonly found along the shoulders of roads and in disturbed land, it is generally considered a weed.

A flagellum is a hairlike appendage that protrudes from certain plant and animal sperm cells, from fungal spores (zoospores), and from a wide range of microorganisms to provide motility. Many protists with flagella are known as flagellates.

Toll-like receptors (TLRs) are a class of proteins that play a key role in the innate immune system. They are single-spanning receptors usually expressed on sentinel cells such as macrophages and dendritic cells, that recognize structurally conserved molecules derived from microbes. Once these microbes have reached physical barriers such as the skin or intestinal tract mucosa, they are recognized by TLRs, which activate immune cell responses. The TLRs include TLR1, TLR2, TLR3, TLR4, TLR5, TLR6, TLR7, TLR8, TLR9, TLR10, TLR11, TLR12, and TLR13. Humans lack genes for TLR11, TLR12 and TLR13 and mice lack a functional gene for TLR10. The receptors TLR1, TLR2, TLR4, TLR5, TLR6, and TLR10 are located on the cell membrane, whereas TLR3, TLR7, TLR8, and TLR9 are located in intracellular vesicles.
Pathogen-associated molecular patterns (PAMPs) are small molecular motifs conserved within a class of microbes, but not present in the host. They are recognized by toll-like receptors (TLRs) and other pattern recognition receptors (PRRs) in both plants and animals. This allows the innate immune system to recognize pathogens and thus, protect the host from infection.
Pattern recognition receptors (PRRs) play a crucial role in the proper function of the innate immune system. PRRs are germline-encoded host sensors, which detect molecules typical for the pathogens. They are proteins expressed mainly by cells of the innate immune system, such as dendritic cells, macrophages, monocytes, neutrophils, as well as by epithelial cells, to identify two classes of molecules: pathogen-associated molecular patterns (PAMPs), which are associated with microbial pathogens, and damage-associated molecular patterns (DAMPs), which are associated with components of host's cells that are released during cell damage or death. They are also called primitive pattern recognition receptors because they evolved before other parts of the immune system, particularly before adaptive immunity. PRRs also mediate the initiation of antigen-specific adaptive immune response and release of inflammatory cytokines.

The innate immune system or nonspecific immune system is one of the two main immunity strategies in vertebrates. The innate immune system is an alternate defense strategy and is the dominant immune system response found in plants, fungi, prokaryotes, and invertebrates.
Systemic acquired resistance (SAR) is a "whole-plant" resistance response that occurs following an earlier localized exposure to a pathogen. SAR is analogous to the innate immune system found in animals, and although there are many shared aspects between the two systems, it is thought to be a result of convergent evolution. The systemic acquired resistance response is dependent on the plant hormone, salicylic acid.

IRAK-4, in the IRAK family, is a protein kinase involved in signaling innate immune responses from Toll-like receptors. It also supports signaling from T-cell receptors. IRAK4 contains domain structures which are similar to those of IRAK1, IRAK2, IRAKM and Pelle. IRAK4 is unique compared to IRAK1, IRAK2 and IRAKM in that it functions upstream of the other IRAKs, but is more similar to Pelle in this trait. IRAK4 has important clinical applications.
The gene-for-gene relationship is a concept in plant pathology that plants and their diseases each have single genes that interact with each other during an infection. It was proposed by Harold Henry Flor who was working with rust (Melampsora lini) of flax (Linum usitatissimum). Flor showed that the inheritance of both resistance in the host and parasite ability to cause disease is controlled by pairs of matching genes. One is a plant gene called the resistance (R) gene. The other is a parasite gene called the avirulence (Avr) gene. Plants producing a specific R gene product are resistant towards a pathogen that produces the corresponding Avr gene product. Gene-for-gene relationships are a widespread and very important aspect of plant disease resistance. Another example can be seen with Lactuca serriola versus Bremia lactucae.

Integrin-linked kinase is an enzyme that in humans is encoded by the ILK gene involved with integrin-mediated signal transduction. Mutations in ILK are associated with cardiomyopathies. It is a 59kDa protein originally identified in a yeast-two hybrid screen with integrin β1 as the bait protein. Since its discovery, ILK has been associated with multiple cellular functions including cell migration, proliferation, and adhesion.

Toll-like receptor 5, also known as TLR5, is a protein which in humans is encoded by the TLR5 gene. It is a member of the toll-like receptor (TLR) family. TLR5 is known to recognize bacterial flagellin from invading mobile bacteria. It has been shown to be involved in the onset of many diseases, including Inflammatory bowel disease due to the high expression of TLR in intestinal lamina propria dendritic cells. Recent studies have also shown that malfunctioning of TLR5 is likely related to rheumatoid arthritis, osteoclastogenesis, and bone loss. Abnormal TLR5 functioning is related to the onset of gastric, cervical, endometrial and ovarian cancers.
Resistance genes (R-Genes) are genes in plant genomes that convey plant disease resistance against pathogens by producing R proteins. The main class of R-genes consist of a nucleotide binding domain (NB) and a leucine rich repeat (LRR) domain(s) and are often referred to as (NB-LRR) R-genes or NLRs. Generally, the NB domain binds either ATP/ADP or GTP/GDP. The LRR domain is often involved in protein-protein interactions as well as ligand binding. NB-LRR R-genes can be further subdivided into toll interleukin 1 receptor (TIR-NB-LRR) and coiled-coil (CC-NB-LRR).
BRI1-associated receptor kinase 1 is an important plant protein that has diverse functions in plant development.
Mitogen-activated protein kinase (MAPK) networks are the pathways and signaling of MAPK, which is a protein kinase that consists of amino acids serine and threonine. MAPK pathways have both a positive and negative regulation in plants. A positive regulation of MAPK networks is to help in assisting with stresses from the environment. A negative regulation of MAPK networks is pertaining to a high quantity of reactive oxygen species (ROS) in the plant.

Leucine-rich repeat receptor like protein kinase are plant cell membrane localized Leucine-rich repeat (LRR) receptor kinase that play critical roles in plant innate immunity. Plants have evolved intricate immunity mechanism to combat against pathogen infection by recognizing Pathogen Associated Molecular Patterns (PAMP) and endogenous Damage Associated Molecular Patterns (DAMP). PEPR 1 considered as the first known DAMP receptor of Arabidopsis.
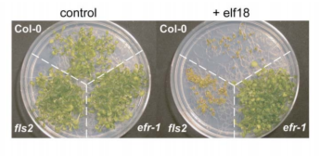
EF-Tu receptor, abbreviated as EFR, is a pattern-recognition receptor (PRR) that binds to the prokaryotic protein EF-Tu in Arabidopsis thaliana. This receptor is an important part of the plant immune system as it allows the plant cells to recognize and bind to EF-Tu, preventing genetic transformation by and protein synthesis in pathogens such as Agrobacterium.

High Affinity K+ transporter HAK5 is a transport protein found on the cell surface membrane of plants under conditions of potassium deprivation. It is believed to act as a symporter for protons and the potassium ion, K+. Firstly discovered in barley, receiving the name of HvHAK1, it was soon after identified in the model plant Arabidopsis thaliana and named HAK5. These transporters belongs to the subgroup I of the KT-HAK-KUP family of plant proteins with obvious homology with both bacterial and fungal transport systems, which experienced a major diversification following land conquest. KT-HAK-KUP transporters are one of four different types of K+ transporter within the cell, but are unique as they do not have a putative pore forming domain like the other three; Shaker channels, KCO channels, HKT transporters. It is activated when the plant is situated in low soil with low potassium concentration, and has been shown to be located in higher concentration in the epidermis and vasculature of K+ deprived plants. By turning on, it increases the plants affinity (uptake) of potassium. Potassium plays a vital role in the plants growth, reproduction, immunity, ion homeostasis, and osmosis, which ensures the plants survival. It is the highest cationic molecule within the plant, accounting for 10% of the plants dry weight, which makes its uptake into the plant important. Each plant species has its own HAK5 transporter that is specific to that species and has different levels of affinity to K+. To operate and activate the HAK5 transporter, the external concentration of K+ must be lower than 10μM and up to 200μM. In Arabidopsis plants, when external potassium concentration is lower than 10μM, it is only HAK5 that is involved with the uptake of K+, then between 10 and 200μM both HAK5 and AKT1 are involved with the uptake of K+. HAK5 is coupled with CBL9/CIPK23 kinase's although the mechanism behind this has not yet been understood.

FLS genes have been discovered to be involved in flagellin reception of bacteria. FLS1 was the original gene discovered shown to correspond with a specific ecotype within Arabidopsis thaliana. Even so, further studies have shown a second FLS gene known as FLS2 that is also associated with flagellin reception. FLS2 and FLS1 are different genes with different responsibilities, but are related genetically. FLS2 has a specific focus in plant defense and is involved in promoting the MAP kinase cascade. Mutations in the FLS2 gene can cause bacterial infection by lack of response to flg22. Therefore,FLS2’s primary focus is association with flg22 while its secondary focus is the involvement of promoting the MAP kinase cascade in plant defense.
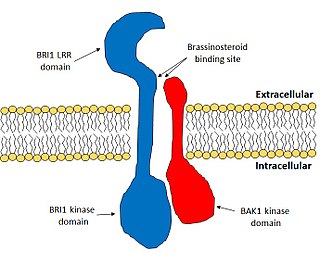
Brassinosteroid insensitive 1 (BRI1) is the major receptor of the plant hormone brassinosteroid. It plays very important roles in plant development, especially in the control of cell elongation and for the tolerance of environmental stresses. BRI1 enhances cell elongation, promotes pollen development, controls vasculature development and promotes chilling and freezing tolerance. BRI1 is one of the most well studied hormone receptors and it acts a model for the study of membrane-bound receptors in plants.