
Ribonucleic acid (RNA) is a polymeric molecule that is essential for most biological functions, either by performing the function itself or by forming a template for the production of proteins. RNA and DNA deoxyribonucleic acid (DNA) are nucleic acids. The nucleic acids constitute one of the four major macromolecules essential for all known forms of life. RNA is assembled as a chain of nucleotides. Cellular organisms use messenger RNA (mRNA) to convey genetic information that directs synthesis of specific proteins. Many viruses encode their genetic information using an RNA genome.

A non-coding RNA (ncRNA) is a functional RNA molecule that is not translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important types of non-coding RNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, exRNAs, scaRNAs and the long ncRNAs such as Xist and HOTAIR.
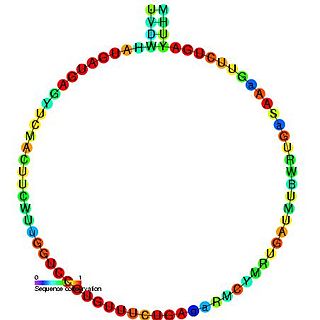
In molecular biology, SNORD18 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, U24 is a member of the C/D class of snoRNA which contain the C (UGAUGA) and D (CUGA) box motifs. C/D box snoRNAs have been shown to act as methylation guides for a number of RNA targets. U24 is encoded within an intron of the gene for ribosomal protein L7a in mammals, chicken and Fugu. The U76/SNORD76 snoRNA is found in an intron of the uRNA host gene (UHG) growth arrest specific 5 (GAS5) transcript gene. snoRNAs Z20 and U76 snoRNAs show clear similarity to U24.
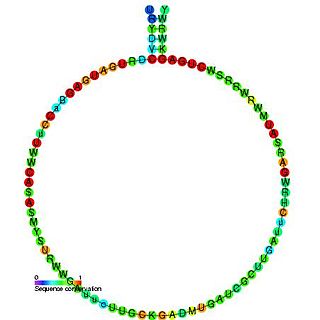
In molecular biology, snoRNA U25 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.
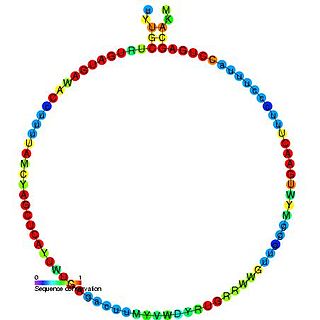
In molecular biology, SNORD29 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, snoRNA U36 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.
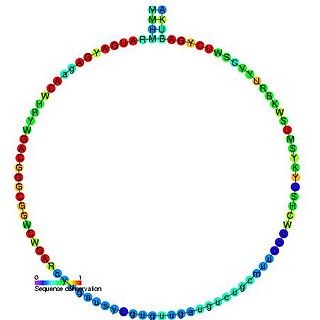
In molecular biology, snoRNA U43 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, snoRNA U50 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, snoRNA U79 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, U81 is a member of the C/D class of snoRNA which contain the C (UGAUGA) and D (CUGA) box motifs. U81 acts as a guanine methylation guide and is found in intron 11 of the gas5 gene in mammals.

In molecular biology, Small Nucleolar RNA SNORD75 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

40S ribosomal protein S30 is a protein that in humans is encoded by the FAU gene.

Long non-coding RNAs are a type of RNA, generally defined as transcripts more than 200 nucleotides that are not translated into protein. This arbitrary limit distinguishes long ncRNAs from small non-coding RNAs, such as microRNAs (miRNAs), small interfering RNAs (siRNAs), Piwi-interacting RNAs (piRNAs), small nucleolar RNAs (snoRNAs), and other short RNAs. Given that some lncRNAs have been reported to have the potential to encode small proteins or micro-peptides, the latest definition of lncRNA is a class of RNA molecules of over 200 nucleotides that have no or limited coding capacity. Long intervening/intergenic noncoding RNAs (lincRNAs) are sequences of lncRNA which do not overlap protein-coding genes.

The Epstein–Barr virus–encoded small RNAs (EBERs) are small non-coding RNAs localized in the nucleus of human cells infected with Epstein–Barr virus (EBV). First discovered in 1981, EBERs are the most abundant RNAs present in infected cells. EBERs interact with several host proteins to form ribonucleoprotein (RNP) complexes. Although a precise function for EBERs remains elusive, roles in transformation and oncogenesis are proposed.
A nested gene is a gene whose entire coding sequence lies within the bounds of a larger external gene. The coding sequence for a nested gene differs greatly from the coding sequence for its external host gene. Typically, nested genes and their host genes encode functionally unrelated proteins, and have different expression patterns in an organism.

HSURs are viral small regulatory RNAs. They are found in Herpesvirus saimiri which is responsible for aggressive T-cell leukemias in primates. They are nuclear RNAs which bind host proteins to form small nuclear ribonucleoproteins (snRNPs). The RNAs are 114–143 nucleotides in length and the HSUR family has been subdivided into HSURs numbered 1 to 7. The function of HSURs has not yet been identified; they do not affect transcription so are thought to act post-transcriptionally, potentially influencing the stability of host mRNAs.
Epstein–Barr virus stable intronic-sequence RNAs (ebv-sisRNAs) are a class of non-coding RNAs generated by repeat introns in the Epstein–Barr virus. After EBERs 1 and 2, ebv-sisRNA-1 is the third most abundant EBV RNA generated during a highly oncogenic form of virus latency. Conservation of ebv-sisRNA sequence and secondary structure between EBV and other herpesviruses suggest shared functions in latent infection.
Small nucleolar RNA host gene 1 is a non-protein coding RNA that in humans is encoded by the SNHG1 gene.

A majority of the human genome is made up of non-protein coding DNA. It infers that such sequences are not commonly employed to encode for a protein. However, even though these regions do not code for protein, they have other functions and carry necessary regulatory information.They can be classified based on the size of the ncRNA. Small noncoding RNA is usually categorized as being under 200 bp in length, whereas long noncoding RNA is greater than 200bp. In addition, they can be categorized by their function within the cell; Infrastructural and Regulatory ncRNAs. Infrastructural ncRNAs seem to have a housekeeping role in translation and splicing and include species such as rRNA, tRNA, snRNA.Regulatory ncRNAs are involved in the modification of other RNAs.


















