Microevolution is the change in allele frequencies that occurs over time within a population. This change is due to four different processes: mutation, selection, gene flow and genetic drift. This change happens over a relatively short amount of time compared to the changes termed macroevolution.

In biology, a hybrid is the offspring resulting from combining the qualities of two organisms of different varieties, species or genera through sexual reproduction. Generally, it means that each cell has genetic material from two different organisms, whereas an individual where some cells are derived from a different organism is called a chimera. Hybrids are not always intermediates between their parents, but can show hybrid vigor, sometimes growing larger or taller than either parent. The concept of a hybrid is interpreted differently in animal and plant breeding, where there is interest in the individual parentage. In genetics, attention is focused on the numbers of chromosomes. In taxonomy, a key question is how closely related the parent species are.

In population genetics, the founder effect is the loss of genetic variation that occurs when a new population is established by a very small number of individuals from a larger population. It was first fully outlined by Ernst Mayr in 1942, using existing theoretical work by those such as Sewall Wright. As a result of the loss of genetic variation, the new population may be distinctively different, both genotypically and phenotypically, from the parent population from which it is derived. In extreme cases, the founder effect is thought to lead to the speciation and subsequent evolution of new species.
A quantitative trait locus (QTL) is a locus that correlates with variation of a quantitative trait in the phenotype of a population of organisms. QTLs are mapped by identifying which molecular markers correlate with an observed trait. This is often an early step in identifying the actual genes that cause the trait variation.
Researchers have investigated the relationship between race and genetics as part of efforts to understand how biology may or may not contribute to human racial categorization. Today, the consensus among scientists is that race is a social construct, and that using it as a proxy for genetic differences among populations is misleading.

Medical genetics is the branch of medicine that involves the diagnosis and management of hereditary disorders. Medical genetics differs from human genetics in that human genetics is a field of scientific research that may or may not apply to medicine, while medical genetics refers to the application of genetics to medical care. For example, research on the causes and inheritance of genetic disorders would be considered within both human genetics and medical genetics, while the diagnosis, management, and counselling people with genetic disorders would be considered part of medical genetics.

In population genetics, an ancestry-informative marker (AIM) is a single-nucleotide polymorphism that exhibits substantially different frequencies between different populations. A set of many AIMs can be used to estimate the proportion of ancestry of an individual derived from each population.

Human genetic variation is the genetic differences in and among populations. There may be multiple variants of any given gene in the human population (alleles), a situation called polymorphism.
David Emil Reich is an American geneticist known for his research into the population genetics of ancient humans, including their migrations and the mixing of populations, discovered by analysis of genome-wide patterns of mutations. He is professor in the department of genetics at the Harvard Medical School, and an associate of the Broad Institute. Reich was highlighted as one of Nature's 10 for his contributions to science in 2015. He received the Dan David Prize in 2017, the NAS Award in Molecular Biology, the Wiley Prize, and the Darwin–Wallace Medal in 2019. In 2021 he was awarded the Massry Prize.
In biology, a cline is a measurable gradient in a single characteristic of a species across its geographical range. Clines usually have a genetic, or phenotypic character. They can show either smooth, continuous gradation in a character, or more abrupt changes in the trait from one geographic region to the next.
Race and health refers to how being identified with a specific race influences health. Race is a complex concept that has changed across chronological eras and depends on both self-identification and social recognition. In the study of race and health, scientists organize people in racial categories depending on different factors such as: phenotype, ancestry, social identity, genetic makeup and lived experience. "Race" and ethnicity often remain undifferentiated in health research.
Population structure is the presence of a systematic difference in allele frequencies between subpopulations. In a randomly mating population, allele frequencies are expected to be roughly similar between groups. However, mating tends to be non-random to some degree, causing structure to arise. For example, a barrier like a river can separate two groups of the same species and make it difficult for potential mates to cross; if a mutation occurs, over many generations it can spread and become common in one subpopulation while being completely absent in the other.
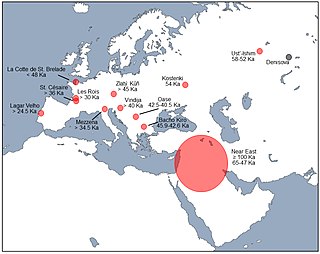
Interbreeding between archaic and modern humans occurred during the Middle Paleolithic and early Upper Paleolithic. The interbreeding happened in several independent events that included Neanderthals and Denisovans, as well as several unidentified hominins.
Genetic studies on Arabs refers to the analyses of the genetics of ethnic Arab people in the Middle East and North Africa. Arabs are genetically diverse as a result of their intermarriage and mixing with indigenous people of the pre-Islamic Middle East and North Africa following the Arab and Islamic expansion. Genetic ancestry components related to the Arabian Peninsula display an increasing frequency pattern from west to east over North Africa. A similar frequency pattern exist across northeastern Africa with decreasing genetic affinities to groups of the Arabian Peninsula along the Nile river valley across Sudan and the more they go south. This genetic cline of admixture is dated to the time of Arab expansion and immigration to North Africa (Maghreb) and northeast Africa.
Mitali Mukerji is a Professor and Head of the Department of Bioscience and Bioengineering, IIT Jodhpur. She was formerly a Chief Scientist at the CSIR Institute of Genomics and Integrative Biology with notable achievement in the field of human genomics and personalized medicine. She is best known for initiating the field of "Ayurgenomics" in partnership with her colleague Dr. Bhavana Prasher under the mentorship of Prof. Samir K. Brahmachari. Ayurgenomics is an innovative study, blending the principles of Ayurveda- the traditional Indian system of medicine- with genomics. Mukerji is also a major contributor in the Indian Genome Variation Consortium, a comprehensive database that is producing "the first genetic landscape of the Indian population", and has been an author in many publications that use IGV databases to study population genomics. Mukerji has done extensive research on hereditary ataxias, and is involved in many other projects related to tracking disease origins and mutational histories. She is the recipient of the prestigious Shanti Swarup Bhatnagar Award in 2010 for her contribution in the field of Medical Sciences.
This glossary of genetics and evolutionary biology is a list of definitions of terms and concepts used in the study of genetics and evolutionary biology, as well as sub-disciplines and related fields, with an emphasis on classical genetics, quantitative genetics, population biology, phylogenetics, speciation, and systematics. Overlapping and related terms can be found in Glossary of cellular and molecular biology, Glossary of ecology, and Glossary of biology.
Genetic studies on Neanderthal ancient DNA became possible in the late 1990s. The Neanderthal genome project, established in 2006, presented the first fully sequenced Neanderthal genome in 2013.
Eukaryote hybrid genomes result from interspecific hybridization, where closely related species mate and produce offspring with admixed genomes. The advent of large-scale genomic sequencing has shown that hybridization is common, and that it may represent an important source of novel variation. Although most interspecific hybrids are sterile or less fit than their parents, some may survive and reproduce, enabling the transfer of adaptive variants across the species boundary, and even result in the formation of novel evolutionary lineages. There are two main variants of hybrid species genomes: allopolyploid, which have one full chromosome set from each parent species, and homoploid, which are a mosaic of the parent species genomes with no increase in chromosome number.
Human genetic clustering refers to patterns of relative genetic similarity among human individuals and populations, as well as the wide range of scientific and statistical methods used to study this aspect of human genetic variation.

The genetic history of Africa summarizes the genetic makeup and population history of African populations in Africa, composed of the overall genetic history, including the regional genetic histories of North Africa, West Africa, East Africa, Central Africa, and Southern Africa, as well as the recent origin of modern humans in Africa. The Sahara served as a trans-regional passageway and place of dwelling for people in Africa during various humid phases and periods throughout the history of Africa.






