Related Research Articles

Arabidopsis thaliana, the thale cress, mouse-ear cress or arabidopsis, is a small flowering plant native to Eurasia and Africa. A. thaliana is considered a weed; it is found along the shoulders of roads and in disturbed land.

Vernalization is the induction of a plant's flowering process by exposure to the prolonged cold of winter, or by an artificial equivalent. After vernalization, plants have acquired the ability to flower, but they may require additional seasonal cues or weeks of growth before they will actually flower. The term is sometimes used to refer to the need of herbal (non-woody) plants for a period of cold dormancy in order to produce new shoots and leaves, but this usage is discouraged.

Self-pollination is a form of pollination in which pollen from the same plant arrives at the stigma of a flower or at the ovule. There are two types of self-pollination: in autogamy, pollen is transferred to the stigma of the same flower; in geitonogamy, pollen is transferred from the anther of one flower to the stigma of another flower on the same flowering plant, or from microsporangium to ovule within a single (monoecious) gymnosperm. Some plants have mechanisms that ensure autogamy, such as flowers that do not open (cleistogamy), or stamens that move to come into contact with the stigma. The term selfing that is often used as a synonym, is not limited to self-pollination, but also applies to other types of self-fertilization.

William Bateson was an English biologist who was the first person to use the term genetics to describe the study of heredity, and the chief populariser of the ideas of Gregor Mendel following their rediscovery in 1900 by Hugo de Vries and Carl Correns. His 1894 book Materials for the Study of Variation was one of the earliest formulations of the new approach to genetics.
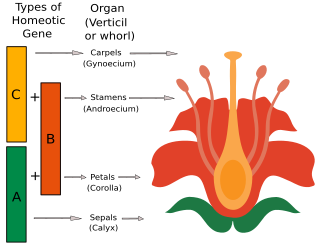
The ABC model of flower development is a scientific model of the process by which flowering plants produce a pattern of gene expression in meristems that leads to the appearance of an organ oriented towards sexual reproduction, a flower. There are three physiological developments that must occur in order for this to take place: firstly, the plant must pass from sexual immaturity into a sexually mature state ; secondly, the transformation of the apical meristem's function from a vegetative meristem into a floral meristem or inflorescence; and finally the growth of the flower's individual organs. The latter phase has been modelled using the ABC model, which aims to describe the biological basis of the process from the perspective of molecular and developmental genetics.
The MADS box is a conserved sequence motif. The genes which contain this motif are called the MADS-box gene family. The MADS box encodes the DNA-binding MADS domain. The MADS domain binds to DNA sequences of high similarity to the motif CC[A/T]6GG termed the CArG-box. MADS-domain proteins are generally transcription factors. The length of the MADS-box reported by various researchers varies somewhat, but typical lengths are in the range of 168 to 180 base pairs, i.e. the encoded MADS domain has a length of 56 to 60 amino acids. There is evidence that the MADS domain evolved from a sequence stretch of a type II topoisomerase in a common ancestor of all extant eukaryotes.
Evolutionary developmental biology (evo-devo) is the study of developmental programs and patterns from an evolutionary perspective. It seeks to understand the various influences shaping the form and nature of life on the planet. Evo-devo arose as a separate branch of science rather recently. An early sign of this occurred in 1999.

Dame Caroline Dean is a British plant scientist working at the John Innes Centre. She is focused on understanding the molecular controls used by plants to seasonally judge when to flower. She is specifically interested in vernalisation — the acceleration of flowering in plants by exposure to periods of prolonged cold. She has also been on the Life Sciences jury for the Infosys Prize from 2018.
RNA polymerase IV is an enzyme that synthesizes small interfering RNA (siRNA) in plants, which silence gene expression. RNAP IV belongs to a family of enzymes that catalyze the process of transcription known as RNA Polymerases, which synthesize RNA from DNA templates. Discovered via phylogenetic studies of land plants, genes of RNAP IV are thought to have resulted from multistep evolution processes that occurred in RNA Polymerase II phylogenies. Such an evolutionary pathway is supported by the fact that RNAP IV is composed of 12 protein subunits that are either similar or identical to RNA polymerase II, and is specific to plant genomes. Via its synthesis of siRNA, RNAP IV is involved in regulation of heterochromatin formation in a process known as RNA directed DNA Methylation (RdDM).
Martin Edward Kreitman is an American geneticist at the University of Chicago, most well known for the McDonald–Kreitman test that is used to infer the amount of adaptive evolution in population genetic studies.

Detlef Weigel is a German American scientist working at the interface of developmental and evolutionary biology.
Michael Webster Bevan is a Professor at the John Innes Centre, Norwich, UK.

Li Jiayang is a Chinese agronomist and geneticist. He is Vice Minister of Agriculture in China and President of the Chinese Academy of Agricultural Sciences (CAAS). He is also Professor and Principal investigator at the Institute of Genetics and Development at the Chinese Academy of Sciences (CAS).
LUX or Phytoclock1 (PCL1) is a gene that codes for LUX ARRHYTHMO, a protein necessary for circadian rhythms in Arabidopsis thaliana. LUX protein associates with Early Flowering 3 (ELF3) and Early Flowering 4 (ELF4) to form the Evening Complex (EC), a core component of the Arabidopsis repressilator model of the plant circadian clock. The LUX protein functions as a transcription factor that negatively regulates Pseudo-Response Regulator 9 (PRR9), a core gene of the Midday Complex, another component of the Arabidopsis repressilator model. LUX is also associated with circadian control of hypocotyl growth factor genes PHYTOCHROME INTERACTING FACTOR 4 (PIF4) and PHYTOCHROME INTERACTING FACTOR 5 (PIF5).
Arabidopsis thaliana is a first class model organism and the single most important species for fundamental research in plant molecular genetics.
Christopher Roland Somerville is a Canadian-American biologist known as a pioneer of Arabidopsis thaliana research. Somerville is currently Professor Emeritus at the University of California, Berkeley and a Program Officer at the Open Philanthropy Project.
A plant genome assembly represents the complete genomic sequence of a plant species, which is assembled into chromosomes and other organelles by using DNA fragments that are obtained from different types of sequencing technology.
DDM1, Decreased DNA Methylation I, is a plant gene that encodes a nucleosome remodeler which facilitates DNA methylation. The DDM1 gene has been described extensively in Arabidopsis thaliana and also in maize. The protein has been described to be similar to the SWI2/SNF2 chromatin remodeling proteins.

RNA-directed DNA methylation (RdDM) is a biological process in which non-coding RNA molecules direct the addition of DNA methylation to specific DNA sequences. The RdDM pathway is unique to plants, although other mechanisms of RNA-directed chromatin modification have also been described in fungi and animals. To date, the RdDM pathway is best characterized within angiosperms, and particularly within the model plant Arabidopsis thaliana. However, conserved RdDM pathway components and associated small RNAs (sRNAs) have also been found in other groups of plants, such as gymnosperms and ferns. The RdDM pathway closely resembles other sRNA pathways, particularly the highly conserved RNAi pathway found in fungi, plants, and animals. Both the RdDM and RNAi pathways produce sRNAs and involve conserved Argonaute, Dicer and RNA-dependent RNA polymerase proteins.
References
- ↑ Krolikowski KA, Victor JL, Wagler TN, Lolle SJ, Pruitt RE (August 2003). "Isolation and characterization of the Arabidopsis organ fusion gene HOTHEAD". Plant J. 35 (4): 501–11. doi: 10.1046/j.1365-313x.2003.01824.x . PMID 12904212.
- ↑ Lolle SJ, Victor JL, Young JM, Pruitt RE (March 2005). "Genome-wide non-mendelian inheritance of extra-genomic information in Arabidopsis". Nature. 434 (7032): 505–9. Bibcode:2005Natur.434..505L. doi:10.1038/nature03380. PMID 15785770. S2CID 1352368.
- ↑ Wade, Nicholas (2005-03-23). "Startling scientists, plant fixes its flawed gene". The New York Times. ISSN 0362-4331 . Retrieved 2016-10-24.
- ↑ Chaudhury, A. (2005). "Hothead healer and extragenomic information". Nature. 437 (7055): E1–E2. Bibcode:2005Natur.437E...1C. doi: 10.1038/nature04062 . PMID 16136082.
- ↑ Comai L, Cartwright RA (2005). "A toxic mutator and selection alternative to the non-Mendelian RNA cache hypothesis for hothead reversion". Plant Cell. 17 (11): 2856–8. doi:10.1105/tpc.105.036293. PMC 1276014 . PMID 16267378. summary Archived 2013-06-21 at the Wayback Machine
- ↑ Krishnaswamy L and Peterson T (2006). "An Alternate Hypothesis to explain the high frequency of "revertants" in Hothead mutants in Arabidopsis". Plant Biology. 9 (1): 30–1. doi:10.1055/s-2006-924563. PMID 17048144.
- ↑ Peng P, Chan SW, Shah GA, Jacobsen SE (September 2006). "Plant genetics: increased outcrossing in hothead mutants". Nature. 443 (7110): E8, discussion E8–9. Bibcode:2006Natur.443E...8P. doi: 10.1038/nature05251 . PMID 17006468.
- ↑ Lolle S. J.; et al. (2006). "Increased outcrossing in hothead mutants (Reply)". Nature. 443 (7110): E8–E9. Bibcode:2006Natur.443E...8L. doi:10.1038/nature05252. S2CID 4425565.
- ↑ Pennisi E (2006). "Genetics. Pollen contamination may explain controversial inheritance". Science. 313 (5795): 1864. doi:10.1126/science.313.5795.1864. PMID 17008492. S2CID 82215542.
- ↑ Mercier R, Jolivet S, Vignard J, et al. (December 2008). "Outcrossing as an explanation of the apparent unconventional genetic behavior of Arabidopsis thaliana hth mutants". Genetics. 180 (4): 2295–7. doi:10.1534/genetics.108.095208. PMC 2600959 . PMID 18845842.