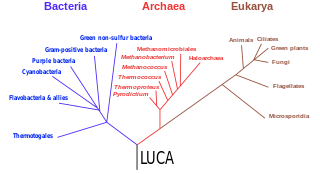
The three-domain system is a taxonomic classification system that groups all cellular life into three domains, namely Archaea, Bacteria and Eukarya, introduced by Carl Woese, Otto Kandler and Mark Wheelis in 1990. The key difference from earlier classifications such as the two-empire system and the five-kingdom classification is the splitting of Archaea from Bacteria as completely different organisms. It has been challenged by the two-domain system that divides organisms into Bacteria and Archaea only, as Eukaryotes are considered as a clade of Archaea.
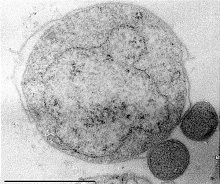
Nanoarchaeum equitans is a species of marine archaea that was discovered in 2002 in a hydrothermal vent off the coast of Iceland on the Kolbeinsey Ridge by Karl Stetter. It has been proposed as the first species in a new phylum, and is the only species within the genus Nanoarchaeum. Strains of this microbe were also found on the Sub-polar Mid Oceanic Ridge, and in the Obsidian Pool in Yellowstone National Park. Since it grows in temperatures approaching boiling, at about 80 °C (176 °F), it is considered to be a thermophile. It grows best in environments with a pH of 6, and a salinity concentration of 2%. Nanoarchaeum appears to be an obligate symbiont on the archaeon Ignicoccus; it must be in contact with the host organism to survive. Nanoarchaeum equitans cannot synthesize lipids but obtains them from its host. Its cells are only 400 nm in diameter, making it the smallest known living organism, and the smallest known archaeon.

The Korarchaeota is a proposed phylum within the Archaea. The name is derived from the Greek noun koros or kore, meaning young man or young woman, and the Greek adjective archaios which means ancient. They are also known as Xenarchaeota. The name is equivalent to Candidatus Korarchaeota, and they go by the name Xenarchaeota or Xenarchaea as well.
The hydrogen hypothesis is a model proposed by William F. Martin and Miklós Müller in 1998 that describes a possible way in which the mitochondrion arose as an endosymbiont within a prokaryotic host in the archaea, giving rise to a symbiotic association of two cells from which the first eukaryotic cell could have arisen (symbiogenesis).

The last universal common ancestor (LUCA) is the hypothesized common ancestral cell from which the three domains of life, the Bacteria, the Archaea, and the Eukarya originated. It is suggested to have been a "cellular organism that had a lipid bilayer and used DNA, RNA, and protein". The LUCA has also been defined as "a hypothetical organism ancestral to all three domains". The LUCA is the point or stage at which the three domains of life diverged from preexisting forms of life. The nature of this point or stage of divergence remains a topic of research.
Viral eukaryogenesis is the hypothesis that the cell nucleus of eukaryotic life forms evolved from a large DNA virus in a form of endosymbiosis within a methanogenic archaeon or a bacterium. The virus later evolved into the eukaryotic nucleus by acquiring genes from the host genome and eventually usurping its role. The hypothesis was first proposed by Philip Bell in 2001 and was further popularized with the discovery of large, complex DNA viruses that are capable of protein biosynthesis.

Loki's Castle is a field of five active hydrothermal vents in the mid-Atlantic Ocean, located at 73 degrees north on the Mid-Atlantic Ridge between Iceland and Svalbard at a depth of 2,352 metres (7,717 ft). They were the most northerly black smoker vents when they were discovered in mid-July 2008.

Archaea is a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotic. Archaea were initially classified as bacteria, receiving the name archaebacteria, but this term has fallen out of use.

The Nitrososphaerota are a phylum of the Archaea proposed in 2008 after the genome of Cenarchaeum symbiosum was sequenced and found to differ significantly from other members of the hyperthermophilic phylum Thermoproteota. Three described species in addition to C. symbiosum are Nitrosopumilus maritimus, Nitrososphaera viennensis, and Nitrososphaera gargensis. The phylum was proposed in 2008 based on phylogenetic data, such as the sequences of these organisms' ribosomal RNA genes, and the presence of a form of type I topoisomerase that was previously thought to be unique to the eukaryotes. This assignment was confirmed by further analysis published in 2010 that examined the genomes of the ammonia-oxidizing archaea Nitrosopumilus maritimus and Nitrososphaera gargensis, concluding that these species form a distinct lineage that includes Cenarchaeum symbiosum. The lipid crenarchaeol has been found only in Nitrososphaerota, making it a potential biomarker for the phylum. Most organisms of this lineage thus far identified are chemolithoautotrophic ammonia-oxidizers and may play important roles in biogeochemical cycles, such as the nitrogen cycle and the carbon cycle. Metagenomic sequencing indicates that they constitute ~1% of the sea surface metagenome across many sites.
The eukaryotes constitute the domain of Eukarya or Eukaryota, organisms whose cells have a membrane-bound nucleus. All animals, plants, fungi, and many unicellular organisms are eukaryotes. They constitute a major group of life forms alongside the two groups of prokaryotes: the Bacteria and the Archaea. Eukaryotes represent a small minority of the number of organisms, but given their generally much larger size, their collective global biomass is much larger than that of prokaryotes.

The eocyte hypothesis in evolutionary biology proposes that the eukaryotes originated from a group of prokaryotes called eocytes. After his team at the University of California, Los Angeles discovered eocytes in 1984, James A. Lake formulated the hypothesis as "eocyte tree" that proposed eukaryotes as part of archaea. Lake hypothesised the tree of life as having only two primary branches: prokaryotes, which include Bacteria and Archaea, and karyotes, that comprise Eukaryotes and eocytes. Parts of this early hypothesis were revived in a newer two-domain system of biological classification which named the primary domains as Archaea and Bacteria.
The "Aigarchaeota" are a proposed archaeal phylum of which the main representative is Caldiarchaeum subterraneum. It is not yet clear if this represents a new phylum or a Nitrososphaerota order, since the genome of Caldiarchaeum subterraneum encodes several Nitrososphaerota-like features. The name "Aigarchaeota" comes from the Greek αυγή, avgí, meaning "dawn" or "aurora", for the intermediate features of hyperthermophilic and mesophilic life during the evolution of its lineage.

"Proteoarchaeota" are a proposed archaeal kingdom thought to be closely related and possibly ancestral to the Eukaryotes.

DPANN is a superphylum of Archaea first proposed in 2013. Many members show novel signs of horizontal gene transfer from other domains of life. They are known as nanoarchaea or ultra-small archaea due to their smaller size (nanometric) compared to other archaea.
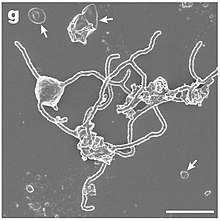
"Candidatus Thorarchaeota", or simply Thorarchaeota, is a phylum within the superphylum Asgard archaea. The Asgard superphylum represents the closest prokaryotic relatives of eukaryotes. Since there is such a close relation between the two different domains, it provides further evidence to the two-domain tree of life theory which states that eukaryotes branched from the archaeal domain. Asgard archaea are single cell marine microbes that contain branch like appendages and have genes that are similar to eukarya. The asgard archaea superphylum is composed of Thorarchaeota, Lokiarchaeota, Odinarchaeota, and Heimdallarchaeota. Thorarchaeota were first identified from the sulfate-methane transition zone in tidewater sediments. Thorarcheota are widely distributed in marine and freshwater sediments.
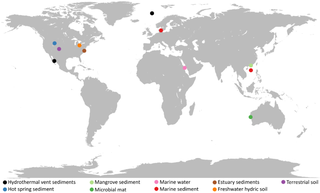
Asgard or Asgardarchaeota is a proposed superphylum consisting of a group of archaea that contain eukaryotic signature proteins. It appears that the eukaryotes, the domain that contains the animals, plants, and fungi, emerged within the Asgard, in a branch containing the Heimdallarchaeota. This supports the two-domain system of classification over the three-domain system.

Marine prokaryotes are marine bacteria and marine archaea. They are defined by their habitat as prokaryotes that live in marine environments, that is, in the saltwater of seas or oceans or the brackish water of coastal estuaries. All cellular life forms can be divided into prokaryotes and eukaryotes. Eukaryotes are organisms whose cells have a nucleus enclosed within membranes, whereas prokaryotes are the organisms that do not have a nucleus enclosed within a membrane. The three-domain system of classifying life adds another division: the prokaryotes are divided into two domains of life, the microscopic bacteria and the microscopic archaea, while everything else, the eukaryotes, become the third domain.

An archaeal virus is a virus that infects and replicates in archaea, a domain of unicellular, prokaryotic organisms. Archaeal viruses, like their hosts, are found worldwide, including in extreme environments inhospitable to most life such as acidic hot springs, highly saline bodies of water, and at the bottom of the ocean. They have been also found in the human body. The first known archaeal virus was described in 1974 and since then, a large diversity of archaeal viruses have been discovered, many possessing unique characteristics not found in other viruses. Little is known about their biological processes, such as how they replicate, but they are believed to have many independent origins, some of which likely predate the last archaeal common ancestor (LACA).
Christa Schleper is a German microbiologist known for her work on the evolution and ecology of Archaea. Schleper is Head of the Department of Functional and Evolutionary Biology at the University of Vienna in Austria.
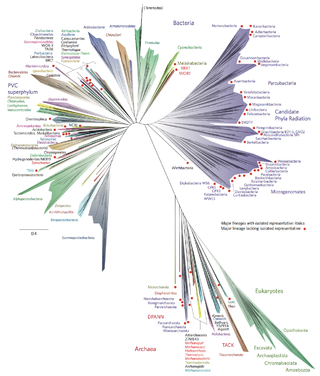
The two-domain system is a biological classification by which all organisms in the tree of life are classified into two domains, Bacteria and Archaea. It emerged from development of knowledge of archaea diversity and challenges to the widely accepted three-domain system that classifies life into Bacteria, Archaea, and Eukarya. It was preceded by the eocyte hypothesis of James A. Lake in the 1980s, which was largely superseded by the three-domain system, due to evidence at the time. Better understanding of archaea, especially of their roles in the origin of eukaryotes through symbiogenesis with bacteria, led to the revival of the eocyte hypothesis in the 2000s. The two-domain system became more widely accepted after the discovery of a large group (superphylum) of archaea called Asgard in 2017, which evidence suggests to be the evolutionary root of eukaryotes, thereby making eukaryotes members of the domain Archaea.