
Signal transduction is the process by which a chemical or physical signal is transmitted through a cell as a series of molecular events, most commonly protein phosphorylation catalyzed by protein kinases, which ultimately results in a cellular response. Proteins responsible for detecting stimuli are generally termed receptors, although in some cases the term sensor is used. The changes elicited by ligand binding in a receptor give rise to a biochemical cascade, which is a chain of biochemical events known as a signaling pathway.
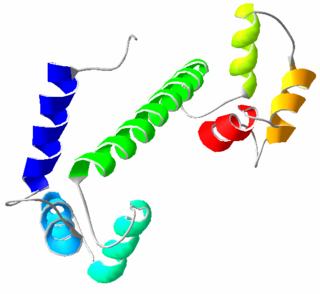
Calmodulin (CaM) (an abbreviation for calcium-modulated protein) is a multifunctional intermediate calcium-binding messenger protein expressed in all eukaryotic cells. It is an intracellular target of the secondary messenger Ca2+, and the binding of Ca2+ is required for the activation of calmodulin. Once bound to Ca2+, calmodulin acts as part of a calcium signal transduction pathway by modifying its interactions with various target proteins such as kinases or phosphatases.

Rhizobia are diazotrophic bacteria that fix nitrogen after becoming established inside the root nodules of legumes (Fabaceae). To express genes for nitrogen fixation, rhizobia require a plant host; they cannot independently fix nitrogen. In general, they are gram negative, motile, non-sporulating rods.

Caspases are a family of protease enzymes playing essential roles in programmed cell death. They are named caspases due to their specific cysteine protease activity – a cysteine in its active site nucleophilically attacks and cleaves a target protein only after an aspartic acid residue. As of 2009, there are 12 confirmed caspases in humans and 10 in mice, carrying out a variety of cellular functions.

Root nodules are found on the roots of plants, primarily legumes, that form a symbiosis with nitrogen-fixing bacteria. Under nitrogen-limiting conditions, capable plants form a symbiotic relationship with a host-specific strain of bacteria known as rhizobia. This process has evolved multiple times within the legumes, as well as in other species found within the Rosid clade. Legume crops include beans, peas, and soybeans.
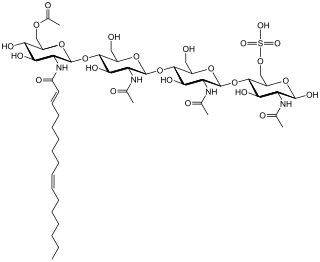
Nod factors, are signaling molecules produced by soil bacteria known as rhizobia in response to flavonoid exudation from plants under nitrogen limited conditions. Nod factors initiate the establishment of a symbiotic relationship between legumes and rhizobia by inducing nodulation. Nod factors produce the differentiation of plant tissue in root hairs into nodules where the bacteria reside and are able to fix nitrogen from the atmosphere for the plant in exchange for photosynthates and the appropriate environment for nitrogen fixation. One of the most important features provided by the plant in this symbiosis is the production of leghemoglobin, which maintains the oxygen concentration low and prevents the inhibition of nitrogenase activity.
Pattern recognition receptors (PRRs) play a crucial role in the proper function of the innate immune system. PRRs are germline-encoded host sensors, which detect molecules typical for the pathogens. They are proteins expressed, mainly, by cells of the innate immune system, such as dendritic cells, macrophages, monocytes, neutrophils and epithelial cells, to identify two classes of molecules: pathogen-associated molecular patterns (PAMPs), which are associated with microbial pathogens, and damage-associated molecular patterns (DAMPs), which are associated with components of host's cells that are released during cell damage or death. They are also called primitive pattern recognition receptors because they evolved before other parts of the immune system, particularly before adaptive immunity. PRRs also mediate the initiation of antigen-specific adaptive immune response and release of inflammatory cytokines.
In biology, cell signaling or cell communication is the ability of a cell to receive, process, and transmit signals with its environment and with itself. Cell signaling is a fundamental property of all cellular life in prokaryotes and eukaryotes. Signals that originate from outside a cell can be physical agents like mechanical pressure, voltage, temperature, light, or chemical signals. Chemical signals can be hydrophobic or hydrophilic. Cell signaling can occur over short or long distances, and as a result can be classified as autocrine, juxtacrine, intracrine, paracrine, or endocrine. Signaling molecules can be synthesized from various biosynthetic pathways and released through passive or active transports, or even from cell damage.

Receptor tyrosine kinases (RTKs) are the high-affinity cell surface receptors for many polypeptide growth factors, cytokines, and hormones. Of the 90 unique tyrosine kinase genes identified in the human genome, 58 encode receptor tyrosine kinase proteins. Receptor tyrosine kinases have been shown not only to be key regulators of normal cellular processes but also to have a critical role in the development and progression of many types of cancer. Mutations in receptor tyrosine kinases lead to activation of a series of signalling cascades which have numerous effects on protein expression. Receptor tyrosine kinases are part of the larger family of protein tyrosine kinases, encompassing the receptor tyrosine kinase proteins which contain a transmembrane domain, as well as the non-receptor tyrosine kinases which do not possess transmembrane domains.

SMAD4, also called SMAD family member 4, Mothers against decapentaplegic homolog 4, or DPC4 is a highly conserved protein present in all metazoans. It belongs to the SMAD family of transcription factor proteins, which act as mediators of TGF-β signal transduction. The TGFβ family of cytokines regulates critical processes during the lifecycle of metazoans, with important roles during embryo development, tissue homeostasis, regeneration, and immune regulation.

The follicle-stimulating hormone receptor or FSH receptor (FSHR) is a transmembrane receptor that interacts with the follicle-stimulating hormone (FSH) and represents a G protein-coupled receptor (GPCR). Its activation is necessary for the hormonal functioning of FSH. FSHRs are found in the ovary, testis, and uterus.

The luteinizing hormone/choriogonadotropin receptor (LHCGR), also lutropin/choriogonadotropin receptor (LCGR) or luteinizing hormone receptor (LHR) is a transmembrane receptor found predominantly in the ovary and testis, but also many extragonadal organs such as the uterus and breasts. The receptor interacts with both luteinizing hormone (LH) and chorionic gonadotropins and represents a G protein-coupled receptor (GPCR). Its activation is necessary for the hormonal functioning during reproduction.

Hypersensitive response (HR) is a mechanism used by plants to prevent the spread of infection by microbial pathogens. HR is characterized by the rapid death of cells in the local region surrounding an infection and it serves to restrict the growth and spread of pathogens to other parts of the plant. It is analogous to the innate immune system found in animals, and commonly precedes a slower systemic response, which ultimately leads to systemic acquired resistance (SAR). HR can be observed in the vast majority of plant species and is induced by a wide range of plant pathogens such as oomycetes, viruses, fungi and even insects.
The gene-for-gene relationship was discovered by Harold Henry Flor who was working with rust of flax. Flor showed that the inheritance of both resistance in the host and parasite ability to cause disease is controlled by pairs of matching genes. One is a plant gene called the resistance (R) gene. The other is a parasite gene called the avirulence (Avr) gene. Plants producing a specific R gene product are resistant towards a pathogen that produces the corresponding Avr gene product. Gene-for-gene relationships are a widespread and very important aspect of plant disease resistance. Another example can be seen with Lactuca serriola versus Bremia lactucae.

Fibroblast growth factor receptor 2 (FGFR2) also known as CD332 is a protein that in humans is encoded by the FGFR2 gene residing on chromosome 10. FGFR2 is a receptor for fibroblast growth factor.

The granulocyte-macrophage colony-stimulating factor receptor also known as CD116, is a receptor for granulocyte-macrophage colony-stimulating factor, which stimulates the production of white blood cells. In contrast to M-CSF and G-CSF which are lineage specific, GM-CSF and its receptor play a role in earlier stages of development. The receptor is primarily located on neutrophils, eosinophils and monocytes/macrophages, it is also on CD34+ progenitor cells (myeloblasts) and precursors for erythroid and megakaryocytic lineages, but only in the beginning of their development.
The ErbB family of proteins contains four receptor tyrosine kinases, structurally related to the epidermal growth factor receptor (EGFR), its first discovered member. In humans, the family includes Her1, Her2, Her3 (ErbB3), and Her4 (ErbB4). The gene symbol, ErbB, is derived from the name of a viral oncogene to which these receptors are homologous: erythroblastic leukemia viral oncogene. Insufficient ErbB signaling in humans is associated with the development of neurodegenerative diseases, such as multiple sclerosis and Alzheimer's disease, while excessive ErbB signaling is associated with the development of a wide variety of types of solid tumor.

C-type lectin domain family 7 member A or Dectin-1 is a protein that in humans is encoded by the CLEC7A gene. CLEC7A is a member of the C-type lectin/C-type lectin-like domain (CTL/CTLD) superfamily. The encoded glycoprotein is a small type II membrane receptor with an extracellular C-type lectin-like domain fold and a cytoplasmic domain with a partial immunoreceptor tyrosine-based activation motif. It functions as a pattern-recognition receptor for a variety of β-1,3-linked and β-1,6-linked glucans from fungi and plants, and in this way plays a role in innate immune response. Expression is found on myeloid dendritic cells, monocytes, macrophages and B cells. Alternate transcriptional splice variants, encoding different isoforms, have been characterized. This gene is closely linked to other CTL/CTLD superfamily members on chromosome 12p13 in the natural killer gene complex region.
The ArpQ Holin Family consists of a single holin-like protein 58 amino acyl residues (aas) in length with 2 transmembrane segments (TMSs). This protein is encoded by the arpQ gene in Enterococcus hirae. While annotated as a holin, it is not functionally characterized.

Paired receptors are pairs or clusters of receptor proteins that bind to extracellular ligands but have opposing activating and inhibitory signaling effects. Traditionally, paired receptors are defined as homologous pairs with similar extracellular domains and different cytoplasmic regions, whose genes are located together in the genome as part of the same gene cluster and which evolved through gene duplication. Homologous paired receptors often, but not always, have a shared ligand in common. More broadly, pairs of receptors have been identified that exhibit paired functional behavior - responding to a shared ligand with opposing intracellular signals - but are not closely homologous or co-located in the genome. Paired receptors are highly expressed in the cells of the immune system, especially natural killer (NK) and myeloid cells, and are involved in immune regulation.















