
The beta sheet is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, Alzheimer's disease and other proteinopathies.

Amyloids are aggregates of proteins characterised by a fibrillar morphology of typically 7–13 nm in diameter, a β-sheet secondary structure and ability to be stained by particular dyes, such as Congo red. In the human body, amyloids have been linked to the development of various diseases. Pathogenic amyloids form when previously healthy proteins lose their normal structure and physiological functions (misfolding) and form fibrous deposits within and around cells. These protein misfolding and deposition processes disrupt the healthy function of tissues and organs.

Amylin, or islet amyloid polypeptide (IAPP), is a 37-residue peptide hormone. It is co-secreted with insulin from the pancreatic β-cells in the ratio of approximately 100:1 (insulin:amylin). Amylin plays a role in glycemic regulation by slowing gastric emptying and promoting satiety, thereby preventing post-prandial spikes in blood glucose levels.

Nuclear magnetic resonance spectroscopy, most commonly known as NMR spectroscopy or magnetic resonance spectroscopy (MRS), is a spectroscopic technique based on re-orientation of atomic nuclei with non-zero nuclear spins in an external magnetic field. This re-orientation occurs with absorption of electromagnetic radiation in the radio frequency region from roughly 4 to 900 MHz, which depends on the isotopic nature of the nucleus and increased proportionally to the strength of the external magnetic field. Notably, the resonance frequency of each NMR-active nucleus depends on its chemical environment. As a result, NMR spectra provide information about individual functional groups present in the sample, as well about connections between nearby nuclei in the same molecule. As the NMR spectra are unique or highly characteristic to individual compounds and functional groups, NMR spectroscopy is one of the most important methods to identify molecular structures, particularly of organic compounds.

Amyloid beta denotes peptides of 36–43 amino acids that are the main component of the amyloid plaques found in the brains of people with Alzheimer's disease. The peptides derive from the amyloid-beta precursor protein (APP), which is cleaved by beta secretase and gamma secretase to yield Aβ in a cholesterol-dependent process and substrate presentation. Aβ molecules can aggregate to form flexible soluble oligomers which may exist in several forms. It is now believed that certain misfolded oligomers can induce other Aβ molecules to also take the misfolded oligomeric form, leading to a chain reaction akin to a prion infection. The oligomers are toxic to nerve cells. The other protein implicated in Alzheimer's disease, tau protein, also forms such prion-like misfolded oligomers, and there is some evidence that misfolded Aβ can induce tau to misfold.

Solid-state NMR (ssNMR) spectroscopy is a technique for characterizing atomic level structure in solid materials e.g. powders, single crystals and amorphous samples and tissues using nuclear magnetic resonance (NMR) spectroscopy. The anisotropic part of many spin interactions are present in solid-state NMR, unlike in solution-state NMR where rapid tumbling motion averages out many of the spin interactions. As a result, solid-state NMR spectra are characterised by larger linewidths than in solution state NMR, which can be utilized to give quantitative information on the molecular structure, conformation and dynamics of the material. Solid-state NMR is often combined with magic angle spinning to remove anisotropic interactions and improve the resolution as well as the sensitivity of the technique.

The Matrix-2 (M2) protein is a proton-selective viroporin, integral in the viral envelope of the influenza A virus. The channel itself is a homotetramer, where the units are helices stabilized by two disulfide bonds, and is activated by low pH. The M2 protein is encoded on the seventh RNA segment together with the M1 protein. Proton conductance by the M2 protein in influenza A is essential for viral replication.
Physalaemin is a tachykinin peptide obtained from the Physalaemus frog, closely related to substance P. Its structure was first elucidated in 1964.
Adriaan "Ad" Bax is a Dutch-American molecular biophysicist. He was born in the Netherlands and is the Chief of the Section on Biophysical NMR Spectroscopy at the National Institutes of Health. He is known for his work on the methodology of biomolecular NMR spectroscopy.

Insulin-degrading enzyme, also known as IDE, is an enzyme.

William (Bill) DeGrado is a professor at the University of California, San Francisco, where he is the Toby Herfindal Presidential Professor of Entrepreneurship and Innovation in the Department of Pharmaceutical Chemistry. As an early pioneer of protein design, he coined the term de novo protein design. He is also active in discovery of small molecule drugs for a variety of human diseases. He is a member of the U.S. National Academy of Sciences (1999), American Academy of Arts & Sciences (1997) and National Academy of Inventors. He also is a scientific cofounder of Pliant therapeutics.
Nucleic acid NMR is the use of nuclear magnetic resonance spectroscopy to obtain information about the structure and dynamics of nucleic acid molecules, such as DNA or RNA. It is useful for molecules of up to 100 nucleotides, and as of 2003, nearly half of all known RNA structures had been determined by NMR spectroscopy.
Robert Tycko is an American biophysicist whose research primarily involves solid state NMR, including the development of new methods and applications to various areas of physics, chemistry, and biology. He is a member of the Laboratory of Chemical Physics in the National Institute of Diabetes and Digestive and Kidney Diseases at the National Institutes of Health in Bethesda, Maryland, USA. He was formerly a member of the Physical Chemistry Research and Materials Chemistry Research departments of AT&T Bell Labs in Murray Hill, New Jersey. His work has contributed to our understanding of geometric phases in spectroscopy, physical properties of fullerenes, skyrmions in 2D electron systems, protein folding, and amyloid fibrils associated with Alzheimer’s disease and prions.

G. Marius Clore MAE, FRSC, FRS is a British-born, Anglo-American molecular biophysicist and structural biologist. He was born in London, U.K. and is a dual U.S./U.K. Citizen. He is a Member of the National Academy of Sciences, a Fellow of the Royal Society, a NIH Distinguished Investigator, and the Chief of the Molecular and Structural Biophysics Section in the Laboratory of Chemical Physics of the National Institute of Diabetes and Digestive and Kidney Diseases at the U.S. National Institutes of Health. He is known for his foundational work in three-dimensional protein and nucleic acid structure determination by biomolecular NMR spectroscopy, for advancing experimental approaches to the study of large macromolecules and their complexes by NMR, and for developing NMR-based methods to study rare conformational states in protein-nucleic acid and protein-protein recognition. Clore's discovery of previously undetectable, functionally significant, rare transient states of macromolecules has yielded fundamental new insights into the mechanisms of important biological processes, and in particular the significance of weak interactions and the mechanisms whereby the opposing constraints of speed and specificity are optimized. Further, Clore's work opens up a new era of pharmacology and drug design as it is now possible to target structures and conformations that have been heretofore unseen.
Marc Baldus is a physicist and professor of NMR spectroscopy at Utrecht University. He is especially known for his work in the field of structural biology using solid-state nuclear magnetic resonance (ssNMR) spectroscopy. He applies ssNMR methods to establish structure-function relationships in complex biomolecular systems including membrane and Amyloid proteins. In addition, he develops cellular NMR methods to study large molecular transport and insertion systems in bacteria as well as signal transduction mechanisms in eukaryotic cells.
The ion channel hypothesis of Alzheimer’s disease (AD), also known as the channel hypothesis or the amyloid beta ion channel hypothesis, is a more recent variant of the amyloid hypothesis of AD, which identifies amyloid beta (Aβ) as the underlying cause of neurotoxicity seen in AD. While the traditional formulation of the amyloid hypothesis pinpoints insoluble, fibrillar aggregates of Aβ as the basis of disruption of calcium ion homeostasis and subsequent apoptosis in AD, the ion channel hypothesis in 1993 introduced the possibility of an ion-channel-forming oligomer of soluble, non-fibrillar Aβ as the cytotoxic species allowing unregulated calcium influx into neurons in AD.
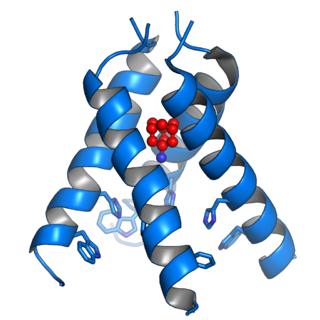
Viroporins are small and usually hydrophobic multifunctional viral proteins that modify cellular membranes, thereby facilitating virus release from infected cells. Viroporins are capable of assembling into oligomeric ion channels or pores in the host cell's membrane, rendering it more permeable and thus facilitating the exit of virions from the cell. Many viroporins also have additional effects on cellular metabolism and homeostasis mediated by protein-protein interactions with host cell proteins. Viroporins are not necessarily essential for viral replication, but do enhance growth rates. They are found in a variety of viral genomes but are particularly common in RNA viruses. Many viruses that cause human disease express viroporins. These viruses include hepatitis C virus, HIV-1, influenza A virus, poliovirus, respiratory syncytial virus, and SARS-CoV.

Hartmut Oschkinat is a German structural biologist and professor for chemistry at the Free University of Berlin. His research focuses on the study of biological systems with solid-state nuclear magnetic resonance.

James J. Chou (周界文) is a Chinese American scientist and Professor of Biological Chemistry and Molecular Pharmacology at the Harvard Medical School. He is known for pioneering the use of Nuclear Magnetic Resonance (NMR) Spectroscopy to reveal the structural details of the membrane regions of cell surface proteins, particularly those of immune receptors and viral membrane proteins.
Suzana K. Straus is a Canadian chemist who is a professor at the University of British Columbia. Her research focuses on host defense peptides (HDPs), as well as protein-protein and protein-ligand interactions.













