Related Research Articles
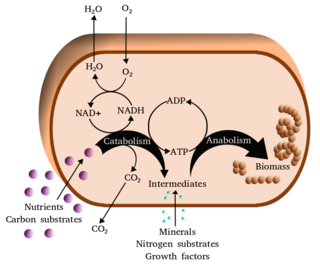
Metabolism is the set of life-sustaining chemical reactions in organisms. The three main functions of metabolism are: the conversion of the energy in food to energy available to run cellular processes; the conversion of food to building blocks of proteins, lipids, nucleic acids, and some carbohydrates; and the elimination of metabolic wastes. These enzyme-catalyzed reactions allow organisms to grow and reproduce, maintain their structures, and respond to their environments. The word metabolism can also refer to the sum of all chemical reactions that occur in living organisms, including digestion and the transportation of substances into and between different cells, in which case the above described set of reactions within the cells is called intermediary metabolism.

In biochemistry, a metabolic pathway is a linked series of chemical reactions occurring within a cell. The reactants, products, and intermediates of an enzymatic reaction are known as metabolites, which are modified by a sequence of chemical reactions catalyzed by enzymes. In most cases of a metabolic pathway, the product of one enzyme acts as the substrate for the next. However, side products are considered waste and removed from the cell.

Pharmacology is the science of drugs and medications, including a substance's origin, composition, pharmacokinetics, pharmacodynamics, therapeutic use, and toxicology. More specifically, it is the study of the interactions that occur between a living organism and chemicals that affect normal or abnormal biochemical function. If substances have medicinal properties, they are considered pharmaceuticals.
A xenobiotic is a chemical substance found within an organism that is not naturally produced or expected to be present within the organism. It can also cover substances that are present in much higher concentrations than are usual. Natural compounds can also become xenobiotics if they are taken up by another organism, such as the uptake of natural human hormones by fish found downstream of sewage treatment plant outfalls, or the chemical defenses produced by some organisms as protection against predators. The term xenobiotic is also used to refer to organs transplanted from one species to another.

Cytochromes P450 are a superfamily of enzymes containing heme as a cofactor that mostly, but not exclusively, function as monooxygenases. In mammals, these proteins oxidize steroids, fatty acids, and xenobiotics, and are important for the clearance of various compounds, as well as for hormone synthesis and breakdown, steroid hormone synthesis, drug metabolism, and the biosynthesis of defensive compounds, fatty acids, and hormones. CYP450 enzymes convert xenobiotics into hydrophilic derivatives, which are more readily excreted. In almost all of the transformations that they catalyze, P450's affect hydroxylation.

Secondary metabolism is a term for pathways and small molecule products of metabolism that are involved in ecological interactions, but are not absolutely required for the survival of the organism. These molecules are sometimes produced by specialized cells, such as laticifers in plants. Secondary metabolites commonly mediate antagonistic interactions, such as competition and predation, as well as mutualistic ones such as pollination and resource mutualisms. Examples of secondary metabolites include antibiotics, pigments and scents. The opposite of secondary metabolites are primary metabolites, which are considered to be essential to the normal growth or development of an organism.

The metabolome refers to the complete set of small-molecule chemicals found within a biological sample. The biological sample can be a cell, a cellular organelle, an organ, a tissue, a tissue extract, a biofluid or an entire organism. The small molecule chemicals found in a given metabolome may include both endogenous metabolites that are naturally produced by an organism as well as exogenous chemicals that are not naturally produced by an organism.
Drug metabolism is the metabolic breakdown of drugs by living organisms, usually through specialized enzymatic systems. More generally, xenobiotic metabolism is the set of metabolic pathways that modify the chemical structure of xenobiotics, which are compounds foreign to an organism's normal biochemistry, such as any drug or poison. These pathways are a form of biotransformation present in all major groups of organisms and are considered to be of ancient origin. These reactions often act to detoxify poisonous compounds. The study of drug metabolism is called pharmacokinetics.

A natural product is a natural compound or substance produced by a living organism—that is, found in nature. In the broadest sense, natural products include any substance produced by life. Natural products can also be prepared by chemical synthesis and have played a central role in the development of the field of organic chemistry by providing challenging synthetic targets. The term natural product has also been extended for commercial purposes to refer to cosmetics, dietary supplements, and foods produced from natural sources without added artificial ingredients.

Metabolic engineering is the practice of optimizing genetic and regulatory processes within cells to increase the cell's production of a certain substance. These processes are chemical networks that use a series of biochemical reactions and enzymes that allow cells to convert raw materials into molecules necessary for the cell's survival. Metabolic engineering specifically seeks to mathematically model these networks, calculate a yield of useful products, and pin point parts of the network that constrain the production of these products. Genetic engineering techniques can then be used to modify the network in order to relieve these constraints. Once again this modified network can be modeled to calculate the new product yield.

Iproniazid is a non-selective, irreversible monoamine oxidase inhibitor (MAOI) of the hydrazine class. It is a xenobiotic that was originally designed to treat tuberculosis, but was later most prominently used as an antidepressant drug. However, it was withdrawn from the market because of its hepatotoxicity. The medical use of iproniazid was discontinued in most of the world in the 1960s, but remained in use in France until its discontinuation in 2015.
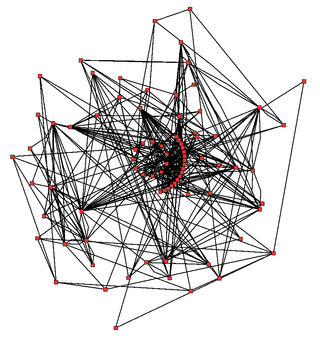
Metabolic network modelling, also known as metabolic network reconstruction or metabolic pathway analysis, allows for an in-depth insight into the molecular mechanisms of a particular organism. In particular, these models correlate the genome with molecular physiology. A reconstruction breaks down metabolic pathways into their respective reactions and enzymes, and analyzes them within the perspective of the entire network. In simplified terms, a reconstruction collects all of the relevant metabolic information of an organism and compiles it in a mathematical model. Validation and analysis of reconstructions can allow identification of key features of metabolism such as growth yield, resource distribution, network robustness, and gene essentiality. This knowledge can then be applied to create novel biotechnology.
Microbial metabolism is the means by which a microbe obtains the energy and nutrients it needs to live and reproduce. Microbes use many different types of metabolic strategies and species can often be differentiated from each other based on metabolic characteristics. The specific metabolic properties of a microbe are the major factors in determining that microbe's ecological niche, and often allow for that microbe to be useful in industrial processes or responsible for biogeochemical cycles.
The non-mevalonate pathway—also appearing as the mevalonate-independent pathway and the 2-C-methyl-D-erythritol 4-phosphate/1-deoxy-D-xylulose 5-phosphate (MEP/DOXP) pathway—is an alternative metabolic pathway for the biosynthesis of the isoprenoid precursors isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP). The currently preferred name for this pathway is the MEP pathway, since MEP is the first committed metabolite on the route to IPP.
Microbial biodegradation is the use of bioremediation and biotransformation methods to harness the naturally occurring ability of microbial xenobiotic metabolism to degrade, transform or accumulate environmental pollutants, including hydrocarbons, polychlorinated biphenyls (PCBs), polyaromatic hydrocarbons (PAHs), heterocyclic compounds, pharmaceutical substances, radionuclides and metals.
The following outline is provided as an overview of and topical guide to cell biology:
Environmental xenobiotics are xenobiotic compounds with a biological activity that are found as pollutants in the natural environment.

Cofactor engineering, a subset of metabolic engineering, is defined as the manipulation of the use of cofactors in an organism’s metabolic pathways. In cofactor engineering, the concentrations of cofactors are changed in order to maximize or minimize metabolic fluxes. This type of engineering can be used to optimize the production of a metabolite product or to increase the efficiency of a metabolic network. The use of engineering single celled organisms to create lucrative chemicals from cheap raw materials is growing, and cofactor engineering can play a crucial role in maximizing production. The field has gained more popularity in the past decade and has several practical applications in chemical manufacturing, bioengineering and pharmaceutical industries.
Pharmacometabolomics, also known as pharmacometabonomics, is a field which stems from metabolomics, the quantification and analysis of metabolites produced by the body. It refers to the direct measurement of metabolites in an individual's bodily fluids, in order to predict or evaluate the metabolism of pharmaceutical compounds, and to better understand the pharmacokinetic profile of a drug. Alternatively, pharmacometabolomics can be applied to measure metabolite levels following the administration of a pharmaceutical compound, in order to monitor the effects of the compound on certain metabolic pathways(pharmacodynamics). This provides detailed mapping of drug effects on metabolism and the pathways that are implicated in mechanism of variation of response to treatment. In addition, the metabolic profile of an individual at baseline (metabotype) provides information about how individuals respond to treatment and highlights heterogeneity within a disease state. All three approaches require the quantification of metabolites found in bodily fluids and tissue, such as blood or urine, and can be used in the assessment of pharmaceutical treatment options for numerous disease states.
Metabolite damage can occur through enzyme promiscuity or spontaneous chemical reactions. Many metabolites are chemically reactive and unstable and can react with other cell components or undergo unwanted modifications. Enzymatically or chemically damaged metabolites are always useless and often toxic. To prevent toxicity that can occur from the accumulation of damaged metabolites, organisms have damage-control systems that:
- Reconvert damaged metabolites to their original, undamaged form
- Convert a potentially harmful metabolite to a benign one
- Prevent damage from happening by limiting the build-up of reactive, but non-damaged metabolites that can lead to harmful products
References
- ↑ Venes, Donald, ed. (1940). Taber's Cyclopedic Medical Dictionary (23 ed.). Philadelphia: F.A. Davis (published 2017). p. 1510. ISBN 9780803659407 . Retrieved 16 March 2020.
metabolite [...] Any product of metabolism.
- ↑ Demain, Arnold L. (December 1980). "Microbial production of primary metabolites". Naturwissenschaften. 67 (12): 582–587. Bibcode:1980NW.....67..582D. doi:10.1007/BF00396537. PMID 7231563. S2CID 21590956.
- ↑ Harris, Edward D. "Biochemical Facts behind the Definition and Properties of Metabolites" (PDF). FDA.gov. United States Food and Drug Administration. Retrieved 28 April 2017.
- ↑ Obach, R. Scott; Esbenshade, Timothy A. (April 2013). "Pharmacologically Active Drug Metabolites: Impact on Drug Discovery and Pharmacotherapy". Pharmacological Reviews. 65 (2): 578–640. doi:10.1124/pr.111.005439. PMID 23406671. S2CID 720243.