
A couple of homologous chromosomes, or homologs, are a set of one maternal and one paternal chromosome that pair up with each other inside a cell during fertilization. Homologs have the same genes in the same loci where they provide points along each chromosome which enable a pair of chromosomes to align correctly with each other before separating during meiosis. This is the basis for Mendelian inheritance which characterizes inheritance patterns of genetic material from an organism to its offspring parent developmental cell at the given time and area.

A nuclease is an enzyme capable of cleaving the phosphodiester bonds between nucleotides of nucleic acids. Nucleases variously effect single and double stranded breaks in their target molecules. In living organisms, they are essential machinery for many aspects of DNA repair. Defects in certain nucleases can cause genetic instability or immunodeficiency. Nucleases are also extensively used in molecular cloning.

DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encodes its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA damage, resulting in tens of thousands of individual molecular lesions per cell per day. Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes. Other lesions induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells after it undergoes mitosis. As a consequence, the DNA repair process is constantly active as it responds to damage in the DNA structure. When normal repair processes fail, and when cellular apoptosis does not occur, irreparable DNA damage may occur, including double-strand breaks and DNA crosslinkages. This can eventually lead to malignant tumors, or cancer as per the two hit hypothesis.
RecQ helicase is a family of helicase enzymes initially found in Escherichia coli that has been shown to be important in genome maintenance. They function through catalyzing the reaction ATP + H2O → ADP + P and thus driving the unwinding of paired DNA and translocating in the 3' to 5' direction. These enzymes can also drive the reaction NTP + H2O → NDP + P to drive the unwinding of either DNA or RNA.

Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids.

Werner syndrome ATP-dependent helicase, also known as DNA helicase, RecQ-like type 3, is an enzyme that in humans is encoded by the WRN gene. WRN is a member of the RecQ Helicase family. Helicase enzymes generally unwind and separate double-stranded DNA. These activities are necessary before DNA can be copied in preparation for cell division. Helicase enzymes are also critical for making a blueprint of a gene for protein production, a process called transcription. Further evidence suggests that Werner protein plays a critical role in repairing DNA. Overall, this protein helps maintain the structure and integrity of a person's DNA.
Mitotic recombination is a type of genetic recombination that may occur in somatic cells during their preparation for mitosis in both sexual and asexual organisms. In asexual organisms, the study of mitotic recombination is one way to understand genetic linkage because it is the only source of recombination within an individual. Additionally, mitotic recombination can result in the expression of recessive alleles in an otherwise heterozygous individual. This expression has important implications for the study of tumorigenesis and lethal recessive alleles. Mitotic homologous recombination occurs mainly between sister chromatids subsequent to replication. Inter-sister homologous recombination is ordinarily genetically silent. During mitosis the incidence of recombination between non-sister homologous chromatids is only about 1% of that between sister chromatids.
Recombinases are genetic recombination enzymes.
The adaptive response is a form of direct DNA repair in E. coli that protects DNA from damage by external agents or by errors during replication. It is initiated against alkylation, particularly methylation, of guanine or thymine nucleotides or phosphate groups on the sugar-phosphate backbone of DNA. Under sustained exposure to low-level treatment with alkylating mutagens, E. coli can adapt to the presence of the mutagen, rendering subsequent treatment with high doses of the same agent less effective.

Serine/threonine-protein kinase ATR, also known as ataxia telangiectasia and Rad3-related protein (ATR) or FRAP-related protein 1 (FRP1), is an enzyme that, in humans, is encoded by the ATR gene. It is a large kinase of about 301.66 kDa. ATR belongs to the phosphatidylinositol 3-kinase-related kinase protein family. ATR is activated in response to single strand breaks, and works with ATM to ensure genome integrity.

Spo11 is a protein that in humans is encoded by the SPO11 gene. Spo11, in a complex with mTopVIB, creates double strand breaks to initiate meiotic recombination. Its active site contains a tyrosine which ligates and dissociates with DNA to promote break formation. One Spo11 protein is involved per strand of DNA, thus two Spo11 proteins are involved in each double stranded break event.

DNA-dependent protein kinase, catalytic subunit, also known as DNA-PKcs, is an enzyme that in humans is encoded by the gene designated as PRKDC or XRCC7. DNA-PKcs belongs to the phosphatidylinositol 3-kinase-related kinase protein family. The DNA-Pkcs protein is a serine/threonine protein kinase consisting of a single polypeptide chain of 4,128 amino acids.

DNA repair and recombination protein RAD54-like is a protein that in humans is encoded by the RAD54L gene.
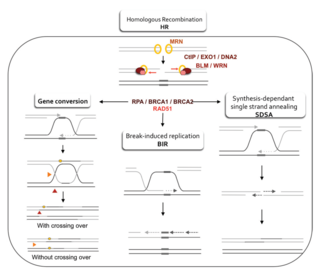
Homology-directed repair (HDR) is a mechanism in cells to repair double-strand DNA lesions. The most common form of HDR is homologous recombination. The HDR mechanism can only be used by the cell when there is a homologous piece of DNA present in the nucleus, mostly in G2 and S phase of the cell cycle. Other examples of homology-directed repair include single-strand annealing and breakage-induced replication. When the homologous DNA is absent, another process called non-homologous end joining (NHEJ) takes place instead.
Microhomology-mediated end joining (MMEJ), also known as alternative nonhomologous end-joining (Alt-NHEJ) is one of the pathways for repairing double-strand breaks in DNA. As reviewed by McVey and Lee, the foremost distinguishing property of MMEJ is the use of microhomologous sequences during the alignment of broken ends before joining, thereby resulting in deletions flanking the original break. MMEJ is frequently associated with chromosome abnormalities such as deletions, translocations, inversions and other complex rearrangements.
Somatic recombination, as opposed to the genetic recombination that occurs in meiosis, is an alteration of the DNA of a somatic cell that is inherited by its daughter cells. The term is usually reserved for large-scale alterations of DNA such as chromosomal translocations and deletions and not applied to point mutations. Somatic recombination occurs physiologically in the assembly of the B cell receptor and T-cell receptor genes, as well as in the class switching of immunoglobulins. Somatic recombination is also important in the process of carcinogenesis.
Genome instability refers to a high frequency of mutations within the genome of a cellular lineage. These mutations can include changes in nucleic acid sequences, chromosomal rearrangements or aneuploidy. Genome instability does occur in bacteria. In multicellular organisms genome instability is central to carcinogenesis, and in humans it is also a factor in some neurodegenerative diseases such as amyotrophic lateral sclerosis or the neuromuscular disease myotonic dystrophy.
DNA damage is an alteration in the chemical structure of DNA, such as a break in a strand of DNA, a nucleobase missing from the backbone of DNA, or a chemically changed base such as 8-OHdG. DNA damage can occur naturally or via environmental factors, but is distinctly different from mutation, although both are types of error in DNA. DNA damage is an abnormal chemical structure in DNA, while a mutation is a change in the sequence of base pairs. DNA damages cause changes in the structure of the genetic material and prevents the replication mechanism from functioning and performing properly. The DNA damage response (DDR) is a complex signal transduction pathway which recognizes when DNA is damaged and initiates the cellular response to the damage.

DNA replication stress refers to the state of a cell whose genome is exposed to various stresses. The events that contribute to replication stress occur during DNA replication, and can result in a stalled replication fork.

A double-strand break repair model refers to the various models of pathways that cells undertake to repair double strand-breaks (DSB). DSB repair is an important cellular process, as the accumulation of unrepaired DSB could lead to chromosomal rearrangements, tumorigenesis or even cell death. In human cells, there are two main DSB repair mechanisms: Homologous recombination (HR) and non-homologous end joining (NHEJ). HR relies on undamaged template DNA as reference to repair the DSB, resulting in the restoration of the original sequence. NHEJ modifies and ligates the damaged ends regardless of homology. In terms of DSB repair pathway choice, most mammalian cells appear to favor NHEJ rather than HR. This is because the employment of HR may lead to gene deletion or amplification in cells which contains repetitive sequences. In terms of repair models in the cell cycle, HR is only possible during the S and G2 phases, while NHEJ can occur throughout whole process. These repair pathways are all regulated by the overarching DNA damage response mechanism. Besides HR and NHEJ, there are also other repair models which exists in cells. Some are categorized under HR, such as synthesis-dependent strain annealing, break-induced replication, and single-strand annealing; while others are an entirely alternate repair model, namely, the pathway microhomology-mediated end joining (MMEJ).













