Related Research Articles

In biochemistry, Michaelis–Menten kinetics is one of the best-known models of enzyme kinetics. It is named after German biochemist Leonor Michaelis and Canadian physician Maud Menten. The model takes the form of an equation describing the rate of enzymatic reactions, by relating reaction rate to , the concentration of a substrate S. Its formula is given by

In biochemistry, the Lineweaver–Burk plot is a graphical representation of the Lineweaver–Burk equation of enzyme kinetics, described by Hans Lineweaver and Dean Burk in 1934. The Lineweaver–Burk plot for inhibited enzymes can be compared to no inhibitor to determine how the inhibitor is competing with the enzyme.

In statistics, nonlinear regression is a form of regression analysis in which observational data are modeled by a function which is a nonlinear combination of the model parameters and depends on one or more independent variables. The data are fitted by a method of successive approximations.

Physiologically based pharmacokinetic (PBPK) modeling is a mathematical modeling technique for predicting the absorption, distribution, metabolism and excretion (ADME) of synthetic or natural chemical substances in humans and other animal species. PBPK modeling is used in pharmaceutical research and drug development, and in health risk assessment for cosmetics or general chemicals.
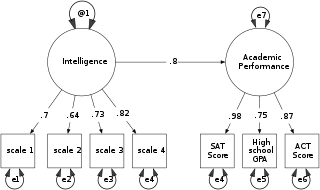
Structural equation modeling (SEM) is a label for a diverse set of methods used by scientists in both experimental and observational research across the sciences, business, and other fields. It is used most in the social and behavioral sciences.
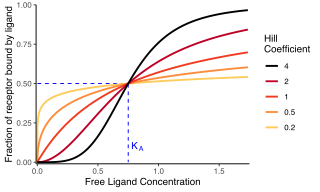
In biochemistry and pharmacology, the Hill equation refers to two closely related equations that reflect the binding of ligands to macromolecules, as a function of the ligand concentration. A ligand is "a substance that forms a complex with a biomolecule to serve a biological purpose", and a macromolecule is a very large molecule, such as a protein, with a complex structure of components. Protein-ligand binding typically changes the structure of the target protein, thereby changing its function in a cell.
In robust statistics, robust regression seeks to overcome some limitations of traditional regression analysis. A regression analysis models the relationship between one or more independent variables and a dependent variable. Standard types of regression, such as ordinary least squares, have favourable properties if their underlying assumptions are true, but can give misleading results otherwise. Robust regression methods are designed to limit the effect that violations of assumptions by the underlying data-generating process have on regression estimates.
Enzyme kinetics is the study of the rates of enzyme-catalysed chemical reactions. In enzyme kinetics, the reaction rate is measured and the effects of varying the conditions of the reaction are investigated. Studying an enzyme's kinetics in this way can reveal the catalytic mechanism of this enzyme, its role in metabolism, how its activity is controlled, and how a drug or a modifier might affect the rate.
In statistics, resampling is the creation of new samples based on one observed sample. Resampling methods are:
- Permutation tests
- Bootstrapping
- Cross validation
Pharmacokinetics, sometimes abbreviated as PK, is a branch of pharmacology dedicated to determining the fate of substances administered to a living organism. The substances of interest include any chemical xenobiotic such as: pharmaceutical drugs, pesticides, food additives, cosmetics, etc. It attempts to analyze chemical metabolism and to discover the fate of a chemical from the moment that it is administered up to the point at which it is completely eliminated from the body. Pharmacokinetics is the study of how an organism affects a drug, whereas pharmacodynamics (PD) is the study of how the drug affects the organism. Both together influence dosing, benefit, and adverse effects, as seen in PK/PD models.
Pharmacometrics is a field of study of the methodology and application of models for disease and pharmacological measurement. It uses mathematical models of biology, pharmacology, disease, and physiology to describe and quantify interactions between xenobiotics and patients, including beneficial effects and adverse effects. It is normally applied to quantify drug, disease and trial information to aid efficient drug development, regulatory decisions and rational drug treatment in patients.
In statistics, best linear unbiased prediction (BLUP) is used in linear mixed models for the estimation of random effects. BLUP was derived by Charles Roy Henderson in 1950 but the term "best linear unbiased predictor" seems not to have been used until 1962. "Best linear unbiased predictions" (BLUPs) of random effects are similar to best linear unbiased estimates (BLUEs) of fixed effects. The distinction arises because it is conventional to talk not about estimating fixed effects but rather about predicting random effects, but the two terms are otherwise equivalent.. However, the equations for the "fixed" effects and for the random effects are different.
Charles Roy Henderson was an American statistician and a pioneer in animal breeding — the application of quantitative methods for the genetic evaluation of domestic livestock. This is critically important because it allows farmers and geneticists to predict whether a crop or animal will have a desired trait, and to what extent the trait will be expressed. He developed mixed model equations to obtain best linear unbiased predictions of breeding values and, in general, any random effect. He invented three methods for the estimation of variance components in unbalanced settings of mixed models, and invented a method for constructing the inverse of Wright's numerator relationship matrix based on a simple list of pedigree information. He, with his Ph.D. student Shayle R. Searle, greatly extended the use of matrix notation in statistics. His methods are widely used by the domestic livestock industry throughout the world and are a cornerstone of linear model theory.
PK/PD modeling is a technique that combines the two classical pharmacologic disciplines of pharmacokinetics and pharmacodynamics. It integrates a pharmacokinetic and a pharmacodynamic model component into one set of mathematical expressions that allows the description of the time course of effect intensity in response to administration of a drug dose. PK/PD modeling is related to the field of pharmacometrics.
Leon Aarons is an Australian pharmacist who researches and teaches in the areas of pharmacodynamics and pharmacokinetics. He lives in the United Kingdom and from 1976 has been a professor of pharmacometrics at the University of Manchester. In the interest of promoting the effective development of drugs, the main focus of his work is optimizing pharmacological models, the design of clinical studies, and data analysis and interpretation in the field of population pharmacokinetics. From 1985 to 2010 Aarons was an editor emeritus of the Journal of Pharmacokinetics and Pharmacodynamics and is a former executive editor of the British Journal of Clinical Pharmacology.
Predictions of surgery duration (SD) are used to schedule planned/elective surgeries so that utilization rate of operating theatres be optimized. An example for a constraint is that a pre-specified tolerance for the percentage of postponed surgeries not be exceeded. The tight linkage between SD prediction and surgery scheduling is the reason that most often scientific research related to scheduling methods addresses also SD predictive methods and vice versa. Durations of surgeries are known to have large variability. Therefore, SD predictive methods attempt, on the one hand, to reduce variability, and on the other employ best available methods to produce SD predictions. The more accurate the predictions, the better the scheduling of surgeries.
Malcolm Rowland FBPhS is Emeritus Professor of Pharmacy, University of Manchester, and Adjunct Professor, University of California San Francisco. His research in pharmacology, has been particularly in physiologically based pharmacokinetics. He has written several textbooks on the subject.

SAAM II is a proprietary and US NIH-funded software that was developed by SAAM Institute in 1997 at the University of Washington in Seattle. SAAM II is a computer program for tracer and pharmacokinetic studies. It is used for compartmental modeling and noncompartmental analysis. Compared to similar software, compartmental models are constructed graphically allowing the quick run of simple systems or the creation of complex structures. The main uses are in metabolic diseases and pharmacokinetics.
Target-mediated drug disposition (TMDD), is the process in which a drug binds with high affinity to its pharmacological target to such an extent that this affects its pharmacokinetic characteristics. Various drug classes can exhibit TMDD, most often these are large compounds but also smaller compounds can exhibit TMDD . A typical TMDD pattern of antibodies displays non-linear clearance and can be seen at concentration ranges that are usually defined as 'mid-to-low'. In this concentration range, the target is partly saturated.
Nonlinear mixed-effects models are a special case of regression analysis for which a range of different software solutions are available. The statistical properties of nonlinear mixed-effects models make direct estimation by a BLUE estimator impossible. Nonlinear mixed effects models are therefore estimated according to Maximum Likelihood principles. Specific estimation methods are applied, such as linearization methods as first-order (FO), first-order conditional (FOCE) or the laplacian (LAPL), approximation methods such as iterative-two stage (ITS), importance sampling (IMP), stochastic approximation estimation (SAEM) or direct sampling. A special case is use of non-parametric approaches. Furthermore, estimation in limited or full Bayesian frameworks is performed using the Metropolis-Hastings or the NUTS algorithms. Some software solutions focus on a single estimation method, others cover a range of estimation methods and/or with interfaces for specific use cases.
References
- ↑ Sheiner, Lewis B.; B. Rosenberg; V.V. Marathe (1977). "Evaluation of Methods for Estimating Population Pharmacokinetic Parameters I. Michaelis-Menten Model: Routine Clinical Pharmacokinetic Data". Journal of Pharmacokinetics and Biopharmaceutics. 5 (5): 445–79. doi:10.1007/BF01061728. PMID 925881. S2CID 28622472.
- ↑ Sheiner, Lewis B.; Stuart L. Beal (1980). "Evaluation of Methods for Estimating Population Pharmacokinetic Parameters I. Michaelis-Menten Model: Routine Clinical Pharmacokinetic Data". Journal of Pharmacokinetics and Biopharmaceutics. 8 (6): 553–71. doi:10.1007/BF01060053. PMID 7229908. S2CID 31174590.
- ↑ Sheiner, Lewis B.; Stuart L. Beal (1981). "Evaluation of Methods for Estimating Population Pharmacokinetic Parameters II. Biexponential Model and Experimental Pharmacokinetic Data". Journal of Pharmacokinetics and Biopharmaceutics. 9 (5): 635–51. doi:10.1007/BF01061030. PMID 7334463. S2CID 2668816.
- ↑ Sheiner, Lewis B.; Stuart L. Beal (1983). "Evaluation of Methods for Estimating Population Pharmacokinetic Parameters III. Monoexponential Model: Routine Clinical Pharmacokinetic Data". Journal of Pharmacokinetics and Biopharmaceutics. 11 (3): 303–19. doi:10.1007/BF01061870. PMID 6644555. S2CID 9308691.
- ↑ Bauer, Robert J. (2019). "NONMEM Tutorial Part I: Description of Commands and Options, With Simple Examples of Population Analysis". CPT: Pharmacometrics & Systems Pharmacology. 8 (8): 525–537. doi:10.1002/psp4.12404. ISSN 2163-8306. PMC 6709426 . PMID 31056834.
- ↑ "PsN :: Home". uupharmacometrics.github.io. Retrieved 2022-05-09.
- ↑ "Wings for NONMEM". wfn.sourceforge.net. Retrieved 2022-05-09.
- ↑ Bauer, Robert J. (2019). "NONMEM Tutorial Part II: Estimation Methods and Advanced Examples". CPT: Pharmacometrics & Systems Pharmacology. 8 (8): 538–556. doi:10.1002/psp4.12422. ISSN 2163-8306. PMC 6709422 . PMID 31044558.