
Virtual Network Computing (VNC) is a graphical desktop-sharing system that uses the Remote Frame Buffer protocol (RFB) to remotely control another computer. It transmits the keyboard and mouse input from one computer to another, relaying the graphical-screen updates, over a network.
The Center for Nanophase Materials Sciences is the first of the five Nanoscale Science Research Centers sponsored by the United States Department of Energy. It is located in Oak Ridge, Tennessee and is a collaborative research facility for the synthesis, characterization, theory/ modeling/ simulation, and design of nanoscale materials. It is co-located with Spallation Neutron Source.
E-Science or eScience is computationally intensive science that is carried out in highly distributed network environments, or science that uses immense data sets that require grid computing; the term sometimes includes technologies that enable distributed collaboration, such as the Access Grid. The term was created by John Taylor, the Director General of the United Kingdom's Office of Science and Technology in 1999 and was used to describe a large funding initiative starting in November 2000. E-science has been more broadly interpreted since then, as "the application of computer technology to the undertaking of modern scientific investigation, including the preparation, experimentation, data collection, results dissemination, and long-term storage and accessibility of all materials generated through the scientific process. These may include data modeling and analysis, electronic/digitized laboratory notebooks, raw and fitted data sets, manuscript production and draft versions, pre-prints, and print and/or electronic publications." In 2014, IEEE eScience Conference Series condensed the definition to "eScience promotes innovation in collaborative, computationally- or data-intensive research across all disciplines, throughout the research lifecycle" in one of the working definitions used by the organizers. E-science encompasses "what is often referred to as big data [which] has revolutionized science... [such as] the Large Hadron Collider (LHC) at CERN... [that] generates around 780 terabytes per year... highly data intensive modern fields of science...that generate large amounts of E-science data include: computational biology, bioinformatics, genomics" and the human digital footprint for the social sciences.
x11vnc is a Virtual Network Computing (VNC) server program. It allows remote access from a remote client to a computer hosting an X Window session and the x11vnc software, continuously polling the X server's frame buffer for changes. This allows the user to control their X11 desktop from a remote computer either on the user's own network, or from over the Internet as if the user were sitting in front of it. x11vnc can also poll non-X11 frame buffer devices, such as webcams or TV tuner cards, iPAQ, Neuros OSD, the Linux console, and the Mac OS X graphics display. x11vnc is part of the LibVNCServer project and is free software available under the GNU General Public License. x11vnc was written by Karl Runge.
United States federal research funders use the term cyberinfrastructure to describe research environments that support advanced data acquisition, data storage, data management, data integration, data mining, data visualization and other computing and information processing services distributed over the Internet beyond the scope of a single institution. In scientific usage, cyberinfrastructure is a technological and sociological solution to the problem of efficiently connecting laboratories, data, computers, and people with the goal of enabling derivation of novel scientific theories and knowledge.
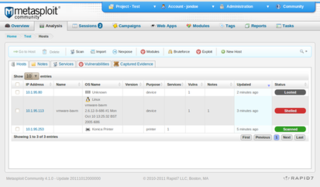
The Metasploit Project is a computer security project that provides information about security vulnerabilities and aids in penetration testing and IDS signature development. It is owned by Boston, Massachusetts-based security company Rapid7.
Rocks Cluster Distribution is a Linux distribution intended for high-performance computing (HPC) clusters. It was started by National Partnership for Advanced Computational Infrastructure and the San Diego Supercomputer Center (SDSC) in 2000. It was initially funded in part by an NSF grant (2000–07), but was funded by the follow-up NSF grant through 2011.
A home server is a computing server located in a private computing residence providing services to other devices inside or outside the household through a home network or the Internet. Such services may include file and printer serving, media center serving, home automation control, web serving, web caching, file sharing and synchronization, video surveillance and digital video recorder, calendar and contact sharing and synchronization, account authentication, and backup services.

A computer cluster is a set of computers that work together so that they can be viewed as a single system. Unlike grid computers, computer clusters have each node set to perform the same task, controlled and scheduled by software.
Web-based simulation (WBS) is the invocation of computer simulation services over the World Wide Web, specifically through a web browser. Increasingly, the web is being looked upon as an environment for providing modeling and simulation applications, and as such, is an emerging area of investigation within the simulation community.

Open Cobalt is a free and open-source software platform for constructing, accessing, and sharing virtual worlds both on local area networks or across the Internet, with no need for centralized servers.
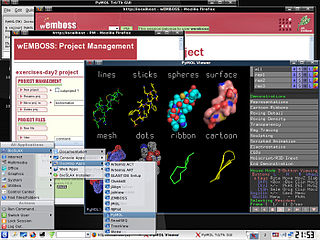
BioSLAX is a Live CD/Live DVD/Live USB comprising a suite of more than 300 bioinformatics tools and application suites. It has been released by the Bioinformatics Resource Unit of the Life Sciences Institute (LSI), National University of Singapore (NUS) and is bootable from any PC that allows a CD/DVD or USB boot option and runs the compressed Slackware flavour of the Linux Operating System (OS), also known as Slax. Slax was created by Tomáš Matějíček in the Czech Republic using the Linux Live Scripts which he also developed. The BioSLAX derivative was created by Mark De Silva, Lim Kuan Siong and Tan Tin Wee.
Techila Distributed Computing Engine is a commercial grid computing software product. It speeds up simulation, analysis and other computational applications by enabling scalability across the IT resources in user's on-premises data center and in the user's own cloud account. Techila Distributed Computing Engine is developed and licensed by Techila Technologies Ltd, a privately held company headquartered in Tampere, Finland. The product is also available as an on-demand solution in Google Cloud Launcher, the online marketplace created and operated by Google. According to IDC, the solution enables organizations to create HPC infrastructure without the major capital investments and operating expenses required by new HPC hardware.
Integrated computational materials engineering (ICME) involves the integration of experimental results, design models, simulations, and other computational data related to a variety of materials used in multiscale engineering and design. Central to the achievement of ICME goals has been the creation of a cyberinfrastructure, a Web-based, collaborative platform which provides the ability to accumulate, organize and disseminate knowledge pertaining to materials science and engineering to facilitate this information being broadly utilized, enhanced, and expanded.

Ninithi is free and open source modelling software that can be used to visualize and analyze carbon materials used in nanotechnology. Users of ninithi can visualize the 3D molecular geometries of graphene/nano-ribbons, carbon nanotubes and fullerenes. Ninithi also provides features to simulate the electronic band structures of graphene and carbon nanotubes. The software was developed by Lanka Software Foundation, in Sri Lanka and released in 2010 under the GPL licence. Ninithi is written in the Java programming language and available for both Microsoft Windows and Linux platforms.

Gerhard Klimeck is a German-American scientist and author in the field of nanotechnology. He is a professor of Electrical and Computer Engineering at Purdue University School of Electrical and Computer Engineering.
Mark S. Lundstrom is an American electrical engineering researcher, educator, and author. He is known for contributions to the theory, modeling, and understanding of semiconductor devices, especially nanoscale transistors, and as the creator of the nanoHUB, a major online resource for nanotechnology. Lundstrom is Don and Carol Scifres Distinguished Professor of Electrical and Computer Engineering and in 2020 served as Acting Dean of the College of Engineering at Purdue University, in West Lafayette, Indiana.
HUBzero is an open source software platform for building websites that support scientific activities.
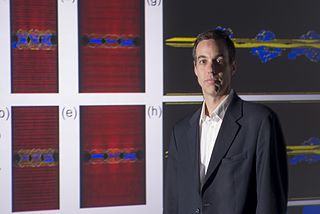
Alejandro Strachan is a scientist in the field of computational materials and a professor of materials engineering at Purdue University. Before joining Purdue University, he was a staff member at Los Alamos National Laboratory.
Science gateways provide access to advanced resources for science and engineering researchers, educators, and students. Through streamlined, online, user-friendly interfaces, gateways combine a variety of cyberinfrastructure (CI) components in support of a community-specific set of tools, applications, and data collections.: In general, these specialized, shared resources are integrated as a Web portal, mobile app, or a suite of applications. Through science gateways, broad communities of researchers can access diverse resources which can save both time and money for themselves and their institutions. As listed below, functions and resources offered by science gateways include shared equipment and instruments, computational services, advanced software applications, collaboration capabilities, data repositories, and networks.








