
A pandemic is an epidemic of an infectious disease that has spread across a large region, for instance multiple continents or worldwide, affecting a substantial number of individuals. Widespread endemic diseases with a stable number of infected individuals such as recurrences of seasonal influenza are generally excluded as they occur simultaneously in large regions of the globe rather than being spread worldwide.

GISAID, the Global Initiative on Sharing All Influenza Data, previously the Global Initiative on Sharing Avian Influenza Data, is a global science initiative established in 2008 to provide access to genomic data of influenza viruses. The database was expanded to include the coronavirus responsible for the COVID-19 pandemic, as well as other pathogens. The database has been described as "the world's largest repository of COVID-19 sequences". GISAID facilitates genomic epidemiology and real-time surveillance to monitor the emergence of new COVID-19 viral strains across the planet.
Marc Van Ranst is a Belgian public health doctor and Professor of Virology at the Katholieke Universiteit Leuven and the Rega Institute for Medical Research. On 1 May 2007, he was appointed as Interministerial comissionar by the Belgian federal government to prepare Belgium for an influenza pandemic.

The Emerging Pathogens Institute (EPI) is an interdisciplinary research institution associated with the University of Florida. The institute focuses on fusing key disciplines to develop outreach, education, and research capabilities designed to preserve the region's health and economy, as well as to prevent or contain new and re-emerging diseases. Researchers within the institute work in more than 30 different countries around the world, with over 250 affiliated faculty members stemming from 11 University of Florida colleges, centers, and institutes. The 90,000-square-foot building includes laboratories and collaborative space for bioinformatics and mathematical modeling.Ebrahim Ahmadpour

Human coronavirus OC43 (HCoV-OC43) is a member of the species Betacoronavirus 1, which infects humans and cattle. The infecting coronavirus is an enveloped, positive-sense, single-stranded RNA virus that enters its host cell by binding to the N-acetyl-9-O-acetylneuraminic acid receptor. OC43 is one of seven coronaviruses known to infect humans. It is one of the viruses responsible for the common cold and may have been responsible for the 1889–1890 pandemic. It has, like other coronaviruses from genus Betacoronavirus, subgenus Embecovirus, an additional shorter spike protein called hemagglutinin-esterase (HE).

Tanja Stadler is a mathematician and professor of computational evolution at the Swiss Federal Institute of Technology. She’s the current president of the Swiss Scientific Advisory Panel COVID-19.

The COVID-19 pandemic, also known as the coronavirus pandemic, is a global pandemic of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The novel virus was first identified in an outbreak in the Chinese city of Wuhan in December 2019. Attempts to contain it there failed, allowing the virus to spread to other areas of Asia and then worldwide in early 2020. The World Health Organization (WHO) declared the outbreak a public health emergency of international concern (PHEIC) on 30 January 2020. The WHO ended its PHEIC declaration on 5 May 2023. As of 18 November 2023, the pandemic had caused 771,820,173 cases and 6,978,162 confirmed deaths, ranking it fifth in the deadliest epidemics and pandemics in history.

Severe acute respiratory syndrome coronavirus 2 (SARS‑CoV‑2) is a strain of coronavirus that causes COVID-19, the respiratory illness responsible for the COVID-19 pandemic. The virus previously had the provisional name 2019 novel coronavirus (2019-nCoV), and has also been called human coronavirus 2019. First identified in the city of Wuhan, Hubei, China, the World Health Organization designated the outbreak a public health emergency of international concern from January 30, 2020, to May 5, 2023. SARS‑CoV‑2 is a positive-sense single-stranded RNA virus that is contagious in humans.
Pandemic prevention is the organization and management of preventive measures against pandemics. Those include measures to reduce causes of new infectious diseases and measures to prevent outbreaks and epidemics from becoming pandemics.

Part of managing an infectious disease outbreak is trying to delay and decrease the epidemic peak, known as flattening the epidemic curve. This decreases the risk of health services being overwhelmed and provides more time for vaccines and treatments to be developed. Non-pharmaceutical interventions that may manage the outbreak include personal preventive measures such as hand hygiene, wearing face masks, and self-quarantine; community measures aimed at physical distancing such as closing schools and cancelling mass gathering events; community engagement to encourage acceptance and participation in such interventions; as well as environmental measures such surface cleaning. It has also been suggested that improving ventilation and managing exposure duration can reduce transmission.

The COVID-19 pandemic has affected animals directly and indirectly. SARS-CoV-2, the virus that causes COVID-19, is zoonotic, which likely to have originated from animals such as bats and pangolins. Human impact on wildlife and animal habitats may be causing such spillover events to become much more likely. The largest incident to date was the culling of 14 to 17 million mink in Denmark after it was discovered that they were infected with a mutant strain of the virus.

The COVID-19 Genomics UK Consortium (COG-UK) was a group of public health agencies and academic institutions in the United Kingdom created in April 2020 to collect, sequence and analyse genomes of SARS-CoV-2 as part of COVID-19 pandemic response. The consortium comprised the UK's four public health agencies, National Health Service organisations, academic partners and the Wellcome Sanger Institute. The consortium was known for first identifying the SARS-CoV-2 Alpha variant in November 2020. As of January 2021, 45% of all SARS-CoV-2 sequences uploaded to the GISAID sequencing database originated from COG-UK.

There are many variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes coronavirus disease 2019 (COVID-19). Some are believed, or have been stated, to be of particular importance due to their potential for increased transmissibility, increased virulence, or reduced effectiveness of vaccines against them. These variants contribute to the continuation of the COVID-19 pandemic.
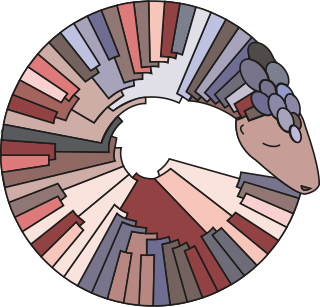
The Phylogenetic Assignment of Named Global Outbreak Lineages (PANGOLIN) is a software tool developed by Dr. Áine O'Toole and members of the Andrew Rambaut laboratory, with an associated web application developed by the Centre for Genomic Pathogen Surveillance in South Cambridgeshire. Its purpose is to implement a dynamic nomenclature to classify genetic lineages for SARS-CoV-2, the virus that causes COVID-19. A user with a full genome sequence of a sample of SARS-CoV-2 can use the tool to submit that sequence, which is then compared with other genome sequences, and assigned the most likely lineage. Single or multiple runs are possible, and the tool can return further information regarding the known history of the assigned lineage. Additionally, it interfaces with Microreact, to show a time sequence of the location of reports of sequenced samples of the same lineage. This latter feature draws on publicly available genomes obtained from the COVID-19 Genomics UK Consortium and from those submitted to GISAID. It is named after the pangolin.

Emma Hodcroft is a British-American molecular epidemiologist at the Institute for Social and Preventive Medicine at the University of Bern. Her research focuses on the phylogenetics of viruses and other pathogens, mapping the spread and evolution of different genetic variants. Hodcroft is a developer on the Nextstrain project, an open science project that tracks the transmission chains of SARS-CoV-2 and other pathogens. In 2020, she originated CoVariants.org, a project tracking SARS-CoV-2 variants.
Jemma Louise Geoghegan is a Scottish-born evolutionary virologist, based at the University of Otago, New Zealand, who specialises in researching emerging infectious diseases and the use of metagenomics to trace the evolution of viruses. As a leader in several government-funded research projects, Geoghegan became the public face of genomic sequencing during New Zealand's response to COVID-19. Her research has contributed to the discussion about the likely cause of COVID-19 and the challenges around predicting pandemics. She was a recipient of the Young Tall Poppy Award in 2017, a Rutherford Discovery Fellowship in 2020, and the 2021 Prime Minister's Emerging Scientist Prize.

The Delta variant (B.1.617.2) was a variant of SARS-CoV-2, the virus that causes COVID-19. It was first detected in India on 5 October 2020. The Delta variant was named on 31 May 2021 and had spread to over 179 countries by 22 November 2021. The World Health Organization (WHO) indicated in June 2021 that the Delta variant was becoming the dominant strain globally.
Trevor Bedford is an American computational virologist at Fred Hutchinson Cancer Research Center.

COVID-19 is predicted to become an endemic disease by many experts. The observed behavior of SARS-CoV-2, the virus that causes COVID-19, suggests it is unlikely it will die out, and the lack of a COVID-19 vaccine that provides long-lasting immunity against infection means it cannot immediately be eradicated; thus, a future transition to an endemic phase appears probable. In an endemic phase, people would continue to become infected and ill, but in relatively stable numbers. Such a transition may take years or decades. Precisely what would constitute an endemic phase is contested.












