α-Ketoglutaric acid is a keto acid.
Pyruvic acid (IUPAC name: 2-oxopropanoic acid, also called acetoic acid) (CH3COCOOH) is the simplest of the alpha-keto acids, with a carboxylic acid and a ketone functional group. Pyruvate, the conjugate base, CH3COCOO−, is an intermediate in several metabolic pathways throughout the cell.

Agrobacterium radiobacter is the causal agent of crown gall disease in over 140 species of eudicots. It is a rod-shaped, Gram-negative soil bacterium. Symptoms are caused by the insertion of a small segment of DNA, from a plasmid into the plant cell, which is incorporated at a semi-random location into the plant genome. Plant genomes can be engineered by use of Agrobacterium for the delivery of sequences hosted in T-DNA binary vectors.

Agrobacterium is a genus of Gram-negative bacteria established by H. J. Conn that uses horizontal gene transfer to cause tumors in plants. Agrobacterium tumefaciens is the most commonly studied species in this genus. Agrobacterium is well known for its ability to transfer DNA between itself and plants, and for this reason it has become an important tool for genetic engineering.

The transfer DNA is the transferred DNA of the tumor-inducing (Ti) plasmid of some species of bacteria such as Agrobacterium tumefaciens and Agrobacterium rhizogenes . The T-DNA is transferred from bacterium into the host plant's nuclear DNA genome. The capability of this specialized tumor-inducing (Ti) plasmid is attributed to two essential regions required for DNA transfer to the host cell. The T-DNA is bordered by 25-base-pair repeats on each end. Transfer is initiated at the right border and terminated at the left border and requires the vir genes of the Ti plasmid.

Malate dehydrogenase (EC 1.1.1.37) (MDH) is an enzyme that reversibly catalyzes the oxidation of malate to oxaloacetate using the reduction of NAD+ to NADH. This reaction is part of many metabolic pathways, including the citric acid cycle. Other malate dehydrogenases, which have other EC numbers and catalyze other reactions oxidizing malate, have qualified names like malate dehydrogenase (NADP+).
In molecular biology, biosynthesis is a multi-step, enzyme-catalyzed process where substrates are converted into more complex products in living organisms. In biosynthesis, simple compounds are modified, converted into other compounds, or joined to form macromolecules. This process often consists of metabolic pathways. Some of these biosynthetic pathways are located within a single cellular organelle, while others involve enzymes that are located within multiple cellular organelles. Examples of these biosynthetic pathways include the production of lipid membrane components and nucleotides. Biosynthesis is usually synonymous with anabolism.

Isocitrate dehydrogenase (IDH) (EC 1.1.1.42) and (EC 1.1.1.41) is an enzyme that catalyzes the oxidative decarboxylation of isocitrate, producing alpha-ketoglutarate (α-ketoglutarate) and CO2. This is a two-step process, which involves oxidation of isocitrate (a secondary alcohol) to oxalosuccinate (a ketone), followed by the decarboxylation of the carboxyl group beta to the ketone, forming alpha-ketoglutarate. In humans, IDH exists in three isoforms: IDH3 catalyzes the third step of the citric acid cycle while converting NAD+ to NADH in the mitochondria. The isoforms IDH1 and IDH2 catalyze the same reaction outside the context of the citric acid cycle and use NADP+ as a cofactor instead of NAD+. They localize to the cytosol as well as the mitochondrion and peroxisome.

A tumour inducing (Ti) plasmid is a plasmid found in pathogenic species of Agrobacterium, including A. tumefaciens, A. rhizogenes, A. rubi and A. vitis.
Shikimic acid, more commonly known as its anionic form shikimate, is a cyclohexene, a cyclitol and a cyclohexanecarboxylic acid. It is an important biochemical metabolite in plants and microorganisms. Its name comes from the Japanese flower shikimi, from which it was first isolated in 1885 by Johan Fredrik Eykman. The elucidation of its structure was made nearly 50 years later.
Opines are low molecular weight compounds found in plant crown gall tumors or hairy root tumors produced by pathogenic bacteria of the genus Agrobacterium and Rhizobium. Opine biosynthesis is catalyzed by specific enzymes encoded by genes contained in a small segment of DNA, which is part of the Ti plasmid or Ri plasmid, inserted by the bacterium into the plant genome. The opines are used by the bacterium as an important energy, carbon and nitrogen source. Each strain of Agrobacterium and Rhizobium induces and catabolizes a specific set of opines, this set typifying the Ti plasmid and Ri plasmid. There are some 30 different opines described so far.

6-Phosphogluconolactonase (EC 3.1.1.31, 6PGL, PGLS, systematic name 6-phospho-D-glucono-1,5-lactone lactonohydrolase) is a cytosolic enzyme found in all organisms that catalyzes the hydrolysis of 6-phosphogluconolactone to 6-phosphogluconic acid in the oxidative phase of the pentose phosphate pathway:
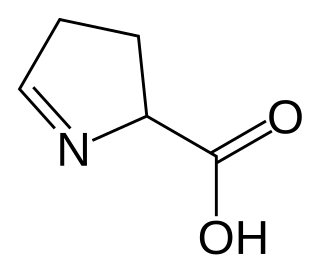
1-Pyrroline-5-carboxylic acid is a cyclic imino acid. Its conjugate base and anion is 1-pyrroline-5-carboxylate (P5C). In solution, P5C is in spontaneous equilibrium with glutamate-5-semialdhyde (GSA).
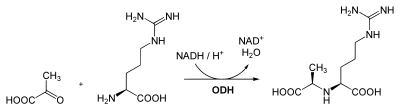
Octopine dehydrogenase (N2-(D-1-carboxyethyl)-L-arginine:NAD+ oxidoreductase, OcDH, ODH) is a dehydrogenase enzyme in the opine dehydrogenase family that helps maintain redox balance under anaerobic conditions. It is found largely in aquatic invertebrates, especially mollusks, sipunculids, and coelenterates, and plays a role analogous to lactate dehydrogenase (found largely in vertebrates) . In the presence of NADH, OcDH catalyzes the reductive condensation of an α-keto acid with an amino acid to form N-carboxyalkyl-amino acids (opines). The purpose of this reaction is to reoxidize glycolytically formed NADH to NAD+, replenishing this important reductant used in glycolysis and allowing for the continued production of ATP in the absence of oxygen.
Delta-1-pyrroline-5-carboxylate synthetase (P5CS) is an enzyme that in humans is encoded by the ALDH18A1 gene. This gene is a member of the aldehyde dehydrogenase family and encodes a bifunctional ATP- and NADPH-dependent mitochondrial enzyme with both gamma-glutamyl kinase and gamma-glutamyl phosphate reductase activities. The encoded protein catalyzes the reduction of glutamate to delta1-pyrroline-5-carboxylate, a critical step in the de novo biosynthesis of proline, ornithine and arginine. Mutations in this gene lead to hyperammonemia, hypoornithinemia, hypocitrullinemia, hypoargininemia and hypoprolinemia and may be associated with neurodegeneration, cataracts and connective tissue diseases. Alternatively spliced transcript variants, encoding different isoforms, have been described for this gene. As reported by Bruno Reversade and colleagues, ALDH18A1 deficiency or dominant-negative mutations in P5CS in humans causes a progeroid disease known as De Barsy Syndrome.

Inosine-5′-monophosphate dehydrogenase (IMPDH) is a purine biosynthetic enzyme that catalyzes the nicotinamide adenine dinucleotide (NAD+)-dependent oxidation of inosine monophosphate (IMP) to xanthosine monophosphate (XMP), the first committed and rate-limiting step towards the de novo biosynthesis of guanine nucleotides from IMP. IMPDH is a regulator of the intracellular guanine nucleotide pool, and is therefore important for DNA and RNA synthesis, signal transduction, energy transfer, glycoprotein synthesis, as well as other process that are involved in cellular proliferation.
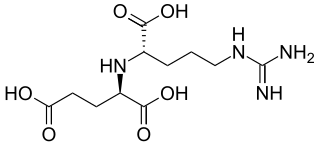
Nopaline is a chemical compound derived from the amino acids glutamic acid and arginine. It is classified as an opine. Ti plasmids are classified on the basis of the different types of opines they produce. These may be nopaline plasmids, octopine plasmids and agropine plasmids. These opines are condensation products of amino acids and keto acids or may be derived from sugars. The opines are used as carbon and nitrogen sources and metabolized by Agrobacterium.

Acetosyringone is a phenolic natural product and a chemical compound related to acetophenone and 2,6-dimethoxyphenol. It was first described in relation to lignan/phenylpropanoid-type phytochemicals, with isolation from a variety of plant sources, in particular, in relation to wounding and other physiologic changes.

In molecular biology, the octopine dehydrogenase family of enzymes act on the CH-NH substrate bond using NAD(+) or NADP(+) as an acceptor. The family includes octopine dehydrogenase EC 1.5.1.11, nopaline dehydrogenase EC 1.5.1.19, lysopine dehydrogenase EC 1.5.1.16 and opine dehydrogenase EC 1.5.1.-. NADPH is the preferred cofactor, but NADH is also used. Octopine dehydrogenase is involved in the reductive condensation of arginine and pyruvic acid to D-octopine.

Bromopyruvic acid is the organic compound with the formula BrCH2COCO2H. This colorless solid is the brominated derivative of pyruvic acid. It bears structural similarity to lactic acid and pyruvic acid. It has been investigated as a metabolic poison and an anticancer agent. Like other α-bromoketones, it is a strong alkylating agent.