An allele is one of two, or more, forms of a given gene variant. For example, the ABO blood grouping is controlled by the ABO gene, which has six common alleles. Nearly every living human's phenotype for the ABO gene is some combination of just these six alleles. An allele is one of two, or more, versions of the same gene at the same place on a chromosome. It can also refer to different sequence variations for a several-hundred base-pair or more region of the genome that codes for a protein. Alleles can come in different extremes of size. At the lowest possible size an allele can be a single nucleotide polymorphism (SNP). At the higher end, it can be up to several thousand base-pairs long. Most alleles result in little or no observable change in the function of the protein the gene codes for.

Genetics is a branch of biology concerned with the study of genes, genetic variation, and heredity in organisms.
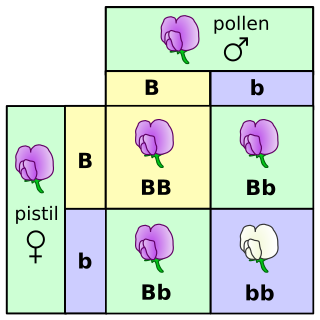
A genotype is an organism’s complete set of genetic material. Often though, genotype is used to refer to a single gene or set of genes, such as the genotype for eye color. The genes take part in determining the characteristics that are observable (phenotype) in an organism, such as hair color, height, etc. An example of a characteristic determined by a genotype is the petal color in a pea plant. The collection of all genetic possibilities for a single trait are called alleles; two alleles for petal color are purple and white.

Heredity, also called inheritance or biological inheritance, is the passing on of traits from parents to their offspring; either through asexual reproduction or sexual reproduction, the offspring cells or organisms acquire the genetic information of their parents. Through heredity, variations between individuals can accumulate and cause species to evolve by natural selection. The study of heredity in biology is genetics.

In biology, a mutation is an alteration in the nucleotide sequence of the genome of an organism, virus, or extrachromosomal DNA. Viral genomes contain either DNA or RNA. Mutations result from errors during DNA or viral replication, mitosis, or meiosis or other types of damage to DNA, which then may undergo error-prone repair, cause an error during other forms of repair, or cause an error during replication. Mutations may also result from insertion or deletion of segments of DNA due to mobile genetic elements.

Wild type (WT) refers to the phenotype of the typical form of a species as it occurs in nature. Originally, the wild type was conceptualized as a product of the standard "normal" allele at a locus, in contrast to that produced by a non-standard, "mutant" allele. "Mutant" alleles can vary to a great extent, and even become the wild type if a genetic shift occurs within the population. Continued advancements in genetic mapping technologies have created a better understanding of how mutations occur and interact with other genes to alter phenotype. It is now appreciated that most or all gene loci exist in a variety of allelic forms, which vary in frequency throughout the geographic range of a species, and that a uniform wild type does not exist. In general, however, the most prevalent allele – i.e., the one with the highest gene frequency – is the one deemed wild type.

In genetics, dominance is the phenomenon of one variant (allele) of a gene on a chromosome masking or overriding the effect of a different variant of the same gene on the other copy of the chromosome. The first variant is termed dominant and the second recessive. This state of having two different variants of the same gene on each chromosome is originally caused by a mutation in one of the genes, either new or inherited. The terms autosomal dominant or autosomal recessive are used to describe gene variants on non-sex chromosomes (autosomes) and their associated traits, while those on sex chromosomes (allosomes) are termed X-linked dominant, X-linked recessive or Y-linked; these have an inheritance and presentation pattern that depends on the sex of both the parent and the child. Since there is only one copy of the Y chromosome, Y-linked traits cannot be dominant nor recessive. Additionally, there are other forms of dominance such as incomplete dominance, in which a gene variant has a partial effect compared to when it is present on both chromosomes, and co-dominance, in which different variants on each chromosome both show their associated traits.
Genetic linkage is the tendency of DNA sequences that are close together on a chromosome to be inherited together during the meiosis phase of sexual reproduction. Two genetic markers that are physically near to each other are unlikely to be separated onto different chromatids during chromosomal crossover, and are therefore said to be more linked than markers that are far apart. In other words, the nearer two genes are on a chromosome, the lower the chance of recombination between them, and the more likely they are to be inherited together. Markers on different chromosomes are perfectly unlinked.

Molecular genetics is a sub-field of biology that addresses how differences in the structures or expression of DNA molecules manifests as variation among organisms. Molecular genetics often applies an "investigative approach" to determine the structure and/or function of genes in an organism's genome using genetic screens. The field of study is based on the merging of several sub-fields in biology: classical Mendelian inheritance, cellular biology, molecular biology, biochemistry, and biotechnology. Researchers search for mutations in a gene or induce mutations in a gene to link a gene sequence to a specific phenotype. Molecular genetics is a powerful methodology for linking mutations to genetic conditions that may aid the search for treatments/cures for various genetics diseases.
A genetic screen or mutagenesis screen is an experimental technique used to identify and select for individuals who possess a phenotype of interest in a mutagenized population. Hence a genetic screen is a type of phenotypic screen. Genetic screens can provide important information on gene function as well as the molecular events that underlie a biological process or pathway. While genome projects have identified an extensive inventory of genes in many different organisms, genetic screens can provide valuable insight as to how those genes function.
A quantitative trait locus (QTL) is a locus that correlates with variation of a quantitative trait in the phenotype of a population of organisms. QTLs are mapped by identifying which molecular markers correlate with an observed trait. This is often an early step in identifying and sequencing the actual genes that cause the trait variation.
Forward genetics is a molecular genetics approach of determining the genetic basis responsible for a phenotype. Forward genetics methods begin with the identification of a phenotype, and finds or creates model organisms that display the characteristic being studied.

Under the law of dominance in genetics, an individual expressing a dominant phenotype could contain either two copies of the dominant allele or one copy of each dominant and recessive allele. By performing a test cross, one can determine whether the individual is homozygous or heterozygous dominant.

In biology, a gene is a basic unit of heredity and a sequence of nucleotides in DNA or RNA that encodes the synthesis of a gene product, either RNA or protein.
Mitotic recombination is a type of genetic recombination that may occur in somatic cells during their preparation for mitosis in both sexual and asexual organisms. In asexual organisms, the study of mitotic recombination is one way to understand genetic linkage because it is the only source of recombination within an individual. Additionally, mitotic recombination can result in the expression of recessive genes in an otherwise heterozygous individual. This expression has important implications for the study of tumorigenesis and lethal recessive genes. Mitotic homologous recombination occurs mainly between sister chromatids subsequent to replication. Inter-sister homologous recombination is ordinarily genetically silent. During mitosis the incidence of recombination between non-sister homologous chromatids is only about 1% of that between sister chromatids.
Lethal alleles are alleles that cause the death of the organism that carries them. They are usually a result of mutations in genes that are essential for growth or development. Lethal alleles may be recessive, dominant, or conditional depending on the gene or genes involved. Lethal alleles can cause death of an organism prenatally or any time after birth, though they commonly manifest early in development.
Uniparental inheritance is a non-mendelian form of inheritance that consists of the transmission of genotypes from one parental type to all progeny. That is, all the genes in offspring will originate from only the mother or only the father. This phenomenon is most commonly observed in eukaryotic organelles such as mitochondria and chloroplasts. This is because such organelles contain their own DNA and are capable of independent mitotic replication that does not endure crossing over with the DNA from another parental type. Although uniparental inheritance is the most common form of inheritance in organelles, there is increased evidence of diversity. Some studies found doubly uniparental inheritance (DUI) and biparental transmission to exist in cells. Evidence suggests that even when there is biparental inheritance, crossing-over doesn't always occur. Furthermore, there is evidence that the form of organelle inheritance varied frequently over time. Uniparental inheritance can be divided into multiple subtypes based on the pathway of inheritance.
Classical genetics is the branch of genetics based solely on visible results of reproductive acts. It is the oldest discipline in the field of genetics, going back to the experiments on Mendelian inheritance by Gregor Mendel who made it possible to identify the basic mechanisms of heredity. Subsequently, these mechanisms have been studied and explained at the molecular level.
Mendelian traits behave according to the model of monogenic or simple gene inheritance in which one gene corresponds to one trait. Discrete traits with simple Mendelian inheritance patterns are relatively rare in nature, and many of the clearest examples in humans cause disorders. Discrete traits found in humans are common examples for teaching genetics.
A human disease modifier gene is a modifier gene that alters expression of a human gene at another locus that in turn causes a genetic disease. Whereas medical genetics has tended to distinguish between monogenic traits, governed by simple, Mendelian inheritance, and quantitative traits, with cumulative, multifactorial causes, increasing evidence suggests that human diseases exist on a continuous spectrum between the two.






