
Deoxyribonucleic acid is a polymer composed of two polynucleotide chains that coil around each other to form a double helix carrying genetic instructions for the development, functioning, growth and reproduction of all known organisms and many viruses. DNA and ribonucleic acid (RNA) are nucleic acids. Alongside proteins, lipids and complex carbohydrates (polysaccharides), nucleic acids are one of the four major types of macromolecules that are essential for all known forms of life.
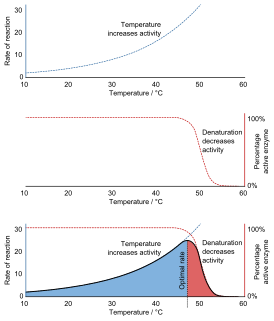
Denaturation is a process in which proteins or nucleic acids lose the quaternary structure, tertiary structure, and secondary structure which is present in their native state, by application of some external stress or compound such as a strong acid or base, a concentrated inorganic salt, an organic solvent, agitation and radiation or heat. If proteins in a living cell are denatured, this results in disruption of cell activity and possibly cell death. Protein denaturation is also a consequence of cell death. Denatured proteins can exhibit a wide range of characteristics, from conformational change and loss of solubility to aggregation due to the exposure of hydrophobic groups. The loss of solubility as a result of denaturation is called coagulation. Denatured proteins lose their 3D structure and therefore cannot function.

Gel electrophoresis is a method for separation and analysis of biomacromolecules and their fragments, based on their size and charge. It is used in clinical chemistry to separate proteins by charge or size and in biochemistry and molecular biology to separate a mixed population of DNA and RNA fragments by length, to estimate the size of DNA and RNA fragments or to separate proteins by charge.

Ribonucleic acid (RNA) is a polymeric molecule essential in various biological roles in coding, decoding, regulation and expression of genes. RNA and deoxyribonucleic acid (DNA) are nucleic acids. Along with lipids, proteins, and carbohydrates, nucleic acids constitute one of the four major macromolecules essential for all known forms of life. Like DNA, RNA is assembled as a chain of nucleotides, but unlike DNA, RNA is found in nature as a single strand folded onto itself, rather than a paired double strand. Cellular organisms use messenger RNA (mRNA) to convey genetic information that directs synthesis of specific proteins. Many viruses encode their genetic information using an RNA genome.
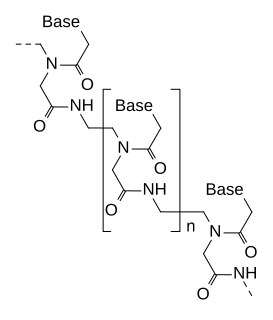
Peptide nucleic acid (PNA) is an artificially synthesized polymer similar to DNA or RNA.
A biosensor is an analytical device, used for the detection of a chemical substance, that combines a biological component with a physicochemical detector. The sensitive biological element, e.g. tissue, microorganisms, organelles, cell receptors, enzymes, antibodies, nucleic acids, etc., is a biologically derived material or biomimetic component that interacts with, binds with, or recognizes the analyte under study. The biologically sensitive elements can also be created by biological engineering. The transducer or the detector element, which transforms one signal into another one, works in a physicochemical way: optical, piezoelectric, electrochemical, electrochemiluminescence etc., resulting from the interaction of the analyte with the biological element, to easily measure and quantify. The biosensor reader device connects with the associated electronics or signal processors that are primarily responsible for the display of the results in a user-friendly way. This sometimes accounts for the most expensive part of the sensor device, however it is possible to generate a user friendly display that includes transducer and sensitive element. The readers are usually custom-designed and manufactured to suit the different working principles of biosensors.
Xenobiology (XB) is a subfield of synthetic biology, the study of synthesizing and manipulating biological devices and systems. The name "xenobiology" derives from the Greek word xenos, which means "stranger, alien". Xenobiology is a form of biology that is not (yet) familiar to science and is not found in nature. In practice, it describes novel biological systems and biochemistries that differ from the canonical DNA–RNA-20 amino acid system. For example, instead of DNA or RNA, XB explores nucleic acid analogues, termed xeno nucleic acid (XNA) as information carriers. It also focuses on an expanded genetic code and the incorporation of non-proteinogenic amino acids into proteins.
Affinity chromatography is a method of separating a biomolecule from a mixture, based on a highly specific macromolecular binding interaction between the biomolecule and another substance. The specific type of binding interaction depends on the biomolecule of interest; antigen and antibody, enzyme and substrate, receptor and ligand, or protein and nucleic acid binding interactions are frequently exploited for isolation of various biomolecules. Affinity chromatography is useful for its high selectivity and resolution of separation, compared to other chromatographic methods.

Streptavidin is a 66.0 (tetramer) kDa protein purified from the bacterium Streptomyces avidinii. Streptavidin homo-tetramers have an extraordinarily high affinity for biotin. With a dissociation constant (Kd) on the order of ≈10−14 mol/L, the binding of biotin to streptavidin is one of the strongest non-covalent interactions known in nature. Streptavidin is used extensively in molecular biology and bionanotechnology due to the streptavidin-biotin complex's resistance to organic solvents, denaturants, detergents, proteolytic enzymes, and extremes of temperature and pH.

Aptamers (APT-uh-murz) are short sequences of artificial DNA or RNA that bind a specific target molecule. Like antibodies, which are used for similar purposes in biotechnology and medicine, they can show strong binding to their target, with little or no off-target binding. To highlight how they are similar to and different from antibodies, aptamers are sometimes called “chemical antibodies.” Aptamers and antibodies can be used in many of the same tasks, but the nucleic acid-based structure of aptamers, which are mostly oligonucleotides, is very different from the amino acid-based structure of antibodies, which are proteins. This difference can make aptamers a better choice than antibodies for some purposes.
Triple-stranded DNA is a DNA structure in which three oligonucleotides wind around each other and form a triple helix. In triple-stranded DNA, the third strand binds to a B-form DNA double helix by forming Hoogsteen base pairs or reversed Hoogsteen hydrogen bonds.
Threose nucleic acid (TNA) is an artificial genetic polymer in which the natural five-carbon ribose sugar found in RNA has been replaced by an unnatural four-carbon threose sugar. Invented by Albert Eschenmoser as part of his quest to explore the chemical etiology of RNA, TNA has become an important synthetic genetic polymer (XNA) due to its ability to efficiently base pair with complementary sequences of DNA and RNA. However, unlike DNA and RNA, TNA is completely refractory to nuclease digestion, making it a promising nucleic acid analog for therapeutic and diagnostic applications.
Targeted drug delivery, sometimes called smart drug delivery, is a method of delivering medication to a patient in a manner that increases the concentration of the medication in some parts of the body relative to others. This means of delivery is largely founded on nanomedicine, which plans to employ nanoparticle-mediated drug delivery in order to combat the downfalls of conventional drug delivery. These nanoparticles would be loaded with drugs and targeted to specific parts of the body where there is solely diseased tissue, thereby avoiding interaction with healthy tissue. The goal of a targeted drug delivery system is to prolong, localize, target and have a protected drug interaction with the diseased tissue. The conventional drug delivery system is the absorption of the drug across a biological membrane, whereas the targeted release system releases the drug in a dosage form. The advantages to the targeted release system is the reduction in the frequency of the dosages taken by the patient, having a more uniform effect of the drug, reduction of drug side-effects, and reduced fluctuation in circulating drug levels. The disadvantage of the system is high cost, which makes productivity more difficult, and the reduced ability to adjust the dosages.
Platinum nanoparticles are usually in the form of a suspension or colloid of nanoparticles of platinum in a fluid, usually water. A colloid is technically defined as a stable dispersion of particles in a fluid medium.

Nucleic acid tertiary structure is the three-dimensional shape of a nucleic acid polymer. RNA and DNA molecules are capable of diverse functions ranging from molecular recognition to catalysis. Such functions require a precise three-dimensional structure. While such structures are diverse and seemingly complex, they are composed of recurring, easily recognizable tertiary structural motifs that serve as molecular building blocks. Some of the most common motifs for RNA and DNA tertiary structure are described below, but this information is based on a limited number of solved structures. Many more tertiary structural motifs will be revealed as new RNA and DNA molecules are structurally characterized.
Hot start PCR is a modified form of conventional polymerase chain reaction (PCR) that reduces the presence of undesired products and primer dimers due to non-specific DNA amplification at room temperatures. Many variations and modifications of the PCR procedure have been developed in order to achieve higher yields; hot start PCR is one of them. Hot start PCR follows the same principles as the conventional PCR - in that it uses DNA polymerase to synthesise DNA from a single stranded template. However, it utilizes additional heating and separation methods, such as inactivating or inhibiting the binding of Taq polymerase and late addition of Taq polymerase, to increase product yield as well as provide a higher specificity and sensitivity. Non-specific binding and priming or formation of primer dimers are minimized by completing the reaction mix after denaturation. Some ways to complete reaction mixes at high temperatures involve modifications that block DNA polymerase activity in low temperatures, use of modified deoxyribonucleotide triphosphates (dNTPs), and the physical addition of one of the essential reagents after denaturation.
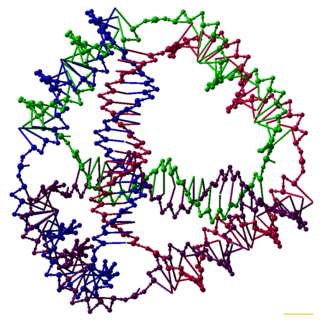
DNA nanotechnology is the design and manufacture of artificial nucleic acid structures for technological uses. In this field, nucleic acids are used as non-biological engineering materials for nanotechnology rather than as the carriers of genetic information in living cells. Researchers in the field have created static structures such as two- and three-dimensional crystal lattices, nanotubes, polyhedra, and arbitrary shapes, and functional devices such as molecular machines and DNA computers. The field is beginning to be used as a tool to solve basic science problems in structural biology and biophysics, including applications in X-ray crystallography and nuclear magnetic resonance spectroscopy of proteins to determine structures. Potential applications in molecular scale electronics and nanomedicine are also being investigated.

Polyvalent DNA gold nanoparticles, now more commonly referred to as spherical nucleic acids, are colloidal gold particles densely modified with short, highly oriented, synthetic DNA strands. They were invented by Chad Mirkin et al. at Northwestern University in 1996. Paul Alivisatos et al. at the University of California, Berkeley introduced a related monovalent structure the same year. Due to the strong interaction between gold and thiols (-SH), the first polyvalent DNA gold nanoparticles were obtained by capping the gold nanoparticles with a dense monolayer of thiol-modified DNA. The dense packing and negative charge of the phosphate backbones of DNA orients it into solution with a footprint that is dependent on factors including the particle size and radius of curvature.

Spherical nucleic acids (SNAs) are nanostructures that consist of a densely packed, highly oriented arrangement of linear nucleic acids in a three-dimensional, spherical geometry. This novel three-dimensional architecture is responsible for many of the SNA's novel chemical, biological, and physical properties that make it useful in biomedicine and materials synthesis. SNAs were first introduced in 1996 by Chad Mirkin’s group at Northwestern University.

Xeno nucleic acids (XNA) are synthetic nucleic acid analogues that have a different sugar backbone than the natural nucleic acids DNA and RNA. As of 2011, at least six types of synthetic sugars have been shown to form nucleic acid backbones that can store and retrieve genetic information. Research is now being done to create synthetic polymerases to transform XNA. The study of its production and application has created a field known as xenobiology.














