
An agar plate is a Petri dish that contains a growth medium solidified with agar, used to culture microorganisms. Sometimes selective compounds are added to influence growth, such as antibiotics.

A cloning vector is a small piece of DNA that can be stably maintained in an organism, and into which a foreign DNA fragment can be inserted for cloning purposes. The cloning vector may be DNA taken from a virus, the cell of a higher organism, or it may be the plasmid of a bacterium. The vector contains features that allow for the convenient insertion of a DNA fragment into the vector or its removal from the vector, for example through the presence of restriction sites. The vector and the foreign DNA may be treated with a restriction enzyme that cuts the DNA, and DNA fragments thus generated contain either blunt ends or overhangs known as sticky ends, and vector DNA and foreign DNA with compatible ends can then be joined together by molecular ligation. After a DNA fragment has been cloned into a cloning vector, it may be further subcloned into another vector designed for more specific use.

The lactose operon is an operon required for the transport and metabolism of lactose in E. coli and many other enteric bacteria. Although glucose is the preferred carbon source for most bacteria, the lac operon allows for the effective digestion of lactose when glucose is not available through the activity of beta-galactosidase. Gene regulation of the lac operon was the first genetic regulatory mechanism to be understood clearly, so it has become a foremost example of prokaryotic gene regulation. It is often discussed in introductory molecular and cellular biology classes for this reason. This lactose metabolism system was used by François Jacob and Jacques Monod to determine how a biological cell knows which enzyme to synthesize. Their work on the lac operon won them the Nobel Prize in Physiology in 1965.

A microbiological culture, or microbial culture, is a method of multiplying microbial organisms by letting them reproduce in predetermined culture medium under controlled laboratory conditions. Microbial cultures are foundational and basic diagnostic methods used as a research tool in molecular biology.
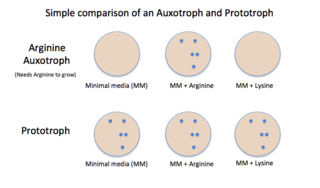
Auxotrophy is the inability of an organism to synthesize a particular organic compound required for its growth. An auxotroph is an organism that displays this characteristic; auxotrophic is the corresponding adjective. Auxotrophy is the opposite of prototrophy, which is characterized by the ability to synthesize all the compounds needed for growth.
Genetics, a discipline of biology, is the science of heredity and variation in living organisms.

A growth medium or culture medium is a solid, liquid, or semi-solid designed to support the growth of a population of microorganisms or cells via the process of cell proliferation or small plants like the moss Physcomitrella patens. Different types of media are used for growing different types of cells.

Chocolate agar (CHOC) or chocolate blood agar (CBA), is a nonselective, enriched growth medium used for isolation of pathogenic bacteria. It is a variant of the blood agar plate, containing red blood cells that have been lysed by slowly heating to 80°C. Chocolate agar is used for growing fastidious respiratory bacteria, such as Haemophilus influenzae and Neisseria meningitidis. In addition, some of these bacteria, most notably H. influenzae, need growth factors such as nicotinamide adenine dinucleotide and hemin, which are inside red blood cells; thus, a prerequisite to growth for these bacteria is the presence of red blood cell lysates. The heat also inactivates enzymes which could otherwise degrade NAD. The agar is named for its color and contains no chocolate products.

Two-hybrid screening is a molecular biology technique used to discover protein–protein interactions (PPIs) and protein–DNA interactions by testing for physical interactions between two proteins or a single protein and a DNA molecule, respectively.
A selectable marker is a gene introduced into a cell, especially a bacterium or to cells in culture, that confers a trait suitable for artificial selection. They are a type of reporter gene used in laboratory microbiology, molecular biology, and genetic engineering to indicate the success of a transfection or other procedure meant to introduce foreign DNA into a cell. Selectable markers are often antibiotic resistance genes. Bacteria that have been subjected to a procedure to introduce foreign DNA are grown on a medium containing an antibiotic, and those bacterial colonies that can grow have successfully taken up and expressed the introduced genetic material. Normally the genes encoding resistance to antibiotics such as ampicillin, chloroamphenicol, tetracycline or kanamycin, etc., are considered useful selectable markers for E. coli.
A colony-forming unit is a unit used in microbiology. It estimates the number of bacteria or fungal cells in a sample which are viable, able to multiply via binary fission under the controlled conditions. Counting with colony-forming units requires culturing the microbes and counts only viable cells, in contrast with microscopic examination which counts all cells, living or dead. The visual appearance of a colony in a cell culture requires significant growth, and when counting colonies it is uncertain if the colony arose from one cell or a group of cells. Expressing results as colony-forming units reflects this uncertainty.
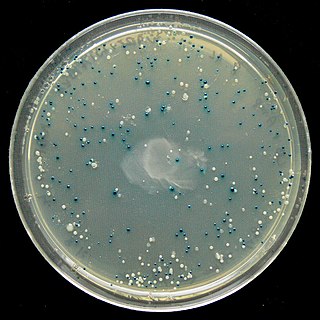
The blue–white screen is a screening technique that allows for the rapid and convenient detection of recombinant bacteria in vector-based molecular cloning experiments. This method of screening is usually performed using a suitable bacterial strain, but other organisms such as yeast may also be used. DNA of transformation is ligated into a vector. The vector is then inserted into a competent host cell viable for transformation, which are then grown in the presence of X-gal. Cells transformed with vectors containing recombinant DNA will produce white colonies; cells transformed with non-recombinant plasmids grow into blue colonies.

The disk diffusion test is a culture-based microbiology assay used in diagnostic and drug discovery laboratories. In diagnostic labs, the assay is used to determine the susceptibility of bacteria isolated from a patient's infection to clinically approved antibiotics. This allows physicians to prescribe the most appropriate antibiotic treatment. In drug discovery labs, especially bioprospecting labs, the assay is used to screen biological material and drug candidates for antibacterial activity. When bioprospecting, the assay can be performed with paired strains of bacteria to achieve dereplication and provisionally identify antibacterial mechanism of action.
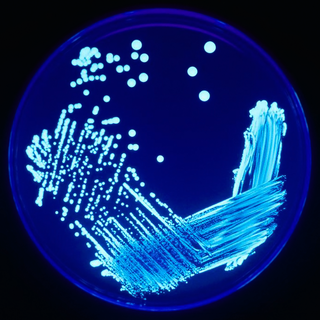
In microbiology, streaking is a technique used to isolate a pure strain from a single species of microorganism, often bacteria. Samples can then be taken from the resulting colonies and a microbiological culture can be grown on a new plate so that the organism can be identified, studied, or tested.

Trypticase soy agar or tryptone soya agar (TSA) and Trypticasesoy broth or tryptone soya broth (TSB) with agar are growth media for the culturing of bacteria. They are general-purpose, nonselective media providing enough nutrients to allow for a wide variety of microorganisms to grow. They are used for a wide range of applications, including culture storage, enumeration of cells (counting), isolation of pure cultures, or simply general culture.

Esther Miriam Zimmer Lederberg was an American microbiologist and a pioneer of bacterial genetics. She discovered the bacterial virus λ and the bacterial fertility factor F, devised the first implementation of replica plating, and furthered the understanding of the transfer of genes between bacteria by specialized transduction.

The 3M Petrifilm plate is an all-in-one plating system made by the Food Safety Division of the 3M Company. They are heavily used in many microbiology-related industries and fields to culture various micro-organisms and are meant to be a more efficient method for detection and enumeration compared to conventional plating techniques. A majority of its use is for the testing of foodstuffs.

Etest is a way of determining antimicrobial sensitivity by placing a strip impregnated with antimicrobials onto an agar plate. A strain of bacterium or fungus will not grow near a concentration of antibiotic or antifungal if it is sensitive. For some microbial and antimicrobial combinations, the results can be used to determine a minimum inhibitory concentration (MIC). Etest is a proprietary system manufactured by bioMérieux. It is a laboratory test used in healthcare settings to help guide physicians by indicating what concentration of antimicrobial could successfully be used to treat patients' infections.
Plate Count Agar (PCA), also called Standard Methods Agar (SMA), is a microbiological growth medium commonly used to assess or to monitor "total" or viable bacterial growth of a sample. PCA is not a selective medium.

The N.Y.C medium or GC medium agar is used for isolating Gonococci.

















