Related Research Articles

An antibiotic is a type of antimicrobial substance active against bacteria. It is the most important type of antibacterial agent for fighting bacterial infections, and antibiotic medications are widely used in the treatment and prevention of such infections. They may either kill or inhibit the growth of bacteria. A limited number of antibiotics also possess antiprotozoal activity. Antibiotics are not effective against viruses such as the common cold or influenza; drugs which inhibit viruses are termed antiviral drugs or antivirals rather than antibiotics.

Antimicrobial resistance (AMR) occurs when microbes evolve mechanisms that protect them from the effects of antimicrobials. Antibiotic resistance is a subset of AMR, that applies specifically to bacteria that become resistant to antibiotics.

Mycoplasma genitalium, is a sexually transmitted, small and pathogenic bacterium that lives on the mucous epithelial cells of the urinary and genital tracts in humans. Medical reports published in 2007 and 2015 state Mgen is becoming increasingly common. Resistance to multiple antibiotics is becoming prevalent, including to azithromycin, which until recently was the most reliable treatment. The bacteria was first isolated from the urogenital tract of humans in 1981, and was eventually identified as a new species of Mycoplasma in 1983. It can cause negative health effects in men and women. It also increases the risk factor for HIV spread with higher occurrences in those previously treated with the azithromycin antibiotics.

Drug resistance is the reduction in effectiveness of a medication such as an antimicrobial or an antineoplastic in treating a disease or condition. The term is used in the context of resistance that pathogens or cancers have "acquired", that is, resistance has evolved. Antimicrobial resistance and antineoplastic resistance challenge clinical care and drive research. When an organism is resistant to more than one drug, it is said to be multidrug-resistant.
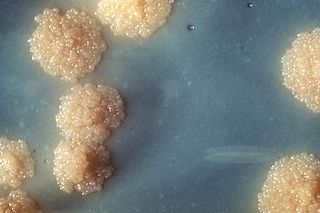
Mycobacterium tuberculosis is a species of pathogenic bacteria in the family Mycobacteriaceae and the causative agent of tuberculosis. First discovered in 1882 by Robert Koch, M. tuberculosis has an unusual, waxy coating on its cell surface primarily due to the presence of mycolic acid. This coating makes the cells impervious to Gram staining, and as a result, M. tuberculosis can appear weakly Gram-positive. Acid-fast stains such as Ziehl–Neelsen, or fluorescent stains such as auramine are used instead to identify M. tuberculosis with a microscope. The physiology of M. tuberculosis is highly aerobic and requires high levels of oxygen. Primarily a pathogen of the mammalian respiratory system, it infects the lungs. The most frequently used diagnostic methods for tuberculosis are the tuberculin skin test, acid-fast stain, culture, and polymerase chain reaction.

Polymyxins are antibiotics. Polymyxins B and E are used in the treatment of Gram-negative bacterial infections. They work mostly by breaking up the bacterial cell membrane. They are part of a broader class of molecules called nonribosomal peptides.

Vancomycin-resistant Staphylococcus aureus (VRSA) are strains of Staphylococcus aureus that have become resistant to the glycopeptide antibiotic vancomycin.

Sequence homology is the biological homology between DNA, RNA, or protein sequences, defined in terms of shared ancestry in the evolutionary history of life. Two segments of DNA can have shared ancestry because of three phenomena: either a speciation event (orthologs), or a duplication event (paralogs), or else a horizontal gene transfer event (xenologs).

Antimicrobial peptides (AMPs), also called host defence peptides (HDPs) are part of the innate immune response found among all classes of life. Fundamental differences exist between prokaryotic and eukaryotic cells that may represent targets for antimicrobial peptides. These peptides are potent, broad spectrum antibiotics which demonstrate potential as novel therapeutic agents. Antimicrobial peptides have been demonstrated to kill Gram negative and Gram positive bacteria, enveloped viruses, fungi and even transformed or cancerous cells. Unlike the majority of conventional antibiotics it appears that antimicrobial peptides frequently destabilize biological membranes, can form transmembrane channels, and may also have the ability to enhance immunity by functioning as immunomodulators.
A gene cassette is a type of mobile genetic element that contains a gene and a recombination site. Each cassette usually contains a single gene and tends to be very small; on the order of 500–1000 base pairs. They may exist incorporated into an integron or freely as circular DNA. Gene cassettes can move around within an organism's genome or be transferred to another organism in the environment via horizontal gene transfer. These cassettes often carry antibiotic resistance genes. An example would be the kanMX cassette which confers kanamycin resistance upon bacteria.

Acinetobacter baumannii is a typically short, almost round, rod-shaped (coccobacillus) Gram-negative bacterium. It is named after the bacteriologist Paul Baumann. It can be an opportunistic pathogen in humans, affecting people with compromised immune systems, and is becoming increasingly important as a hospital-derived (nosocomial) infection. While other species of the genus Acinetobacter are often found in soil samples, it is almost exclusively isolated from hospital environments. Although occasionally it has been found in environmental soil and water samples, its natural habitat is still not known.
The resistome has been used to describe to two similar yet separate concepts:

Fosfomycin, sold under the brand name Berny among others, is an antibiotic primarily used to treat lower UTI. It is not indicated for kidney infections. Occasionally it is used for prostate infections. It is generally taken by mouth.

Beta defensins are a family of mammalian defensins. The beta defensins are antimicrobial peptides implicated in the resistance of epithelial surfaces to microbial colonization.
mecA is a gene found in bacterial cells which allows them to be resistant to antibiotics such as methicillin, penicillin and other penicillin-like antibiotics.
Mustard is a database that tracks Antimicrobial Resistance Determinants (ARDs). The method by which it tracks ARDs is using their own method adapted from Protein Homology Modelling called Pairwise Comparative Modelling (PCM), which increase specificity protein prediction, especially for distantly related protein homologues. Using PCM, 6095 ARDs from 20 families in the human gut microbiota. Antibiotic resistance databases used were ResFinder, ARG-ANNOT, the now defunct Lahey Clinic, Marilyn Roberts website for tetracycline and macrolide resistance genes and metagenomics.
FARME also known as Functional Antibiotic Resistance Metagenomic Element is a database that compiles publicly available DNA elements and predicted proteins that confer antibiotic resistance, regulatory elements and mobile genetic elements. It is the first database to focus on functional metagenomics. This allows the database to understand 99% of bacteria which cannot be cultured, the relationship between environmental antibiotic resistance sequences and antibiotic genes derived from cultured isolates. This information was derived from 20 metagenomics projects from GenBank. Also from GenBank are the protein sequence predictions and annotations.
MUBII-TB-DB is a database that focuses on tuberculosis antibiotic resistance genes. It is a highly structured, text-based database focusing on Mycobacterium tuberculosis at seven different mutation loci: rpoB, pncA, katG; mabA(fabG1)-inhA, gyrA, gyrB, and rrs. MUBII analyzes the query using two parallel strategies: 1). A BLAST search against previously mutated sequences. 2). Alignment of query sequences with wild-type sequences. MUBII outputs graphs of alignments and description of the mutation and therapeutic significance. Therapeutically relevant mutations are tagged as "High-Confident" based on the criteria set by Sandgren et al. MUBII-TB-DB provides a platform that is easy to use for even users that are not trained in bioinformatics.
The SARG database also known as Structured Antibiotic Resistance Gene database is a collection of antimicrobial resistance genes. The hierarchical structure of the database is clear to be 1) Type: antibiotic type 2) Subtype: genotype 3) Sequence: reference sequence. The SARG database helps in quick survey of antimicrobial resistance genes from environmental samples. The database was initially integrated from ARDB and Comprehensive Antibiotic Resistance Database, followed by hand curation including removing non-ARG sequences, removing redundant sequences and SNP sequences. Other sources include NCBI nr database and published papers.
The AMRFinderPlus tool from the National Center for Biotechnology Information (NCBI) is a bioinformatic tool that allows users to identify antimicrobial resistance determinants, stress response, and virulence genes in bacterial genomes. This tool's development began in 2018 and is still underway. The National Institutes of Health funds the development of the software and the databases it uses.
References
- 1 2 Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F. M.; Larsen, M. V. (2012-11-01). "Identification of acquired antimicrobial resistance genes". Journal of Antimicrobial Chemotherapy. 67 (11): 2640–2644. doi:10.1093/jac/dks261. ISSN 0305-7453. PMC 3468078 . PMID 22782487.
- ↑ RATHER, P (1998). "Origins of the aminoglycoside modifying enzymes". Drug Resistance Updates. 1 (5): 285–291. doi:10.1016/s1368-7646(98)80044-7. ISSN 1368-7646. PMID 17092809.
- ↑ van Hoek, Angela H. A. M.; Mevius, Dik; Guerra, Beatriz; Mullany, Peter; Roberts, Adam Paul; Aarts, Henk J. M. (2011). "Acquired Antibiotic Resistance Genes: An Overview". Frontiers in Microbiology. 2: 203. doi: 10.3389/fmicb.2011.00203 . ISSN 1664-302X. PMC 3202223 . PMID 22046172.