The periplasm is a concentrated gel-like matrix in the space between the inner cytoplasmic membrane and the bacterial outer membrane called the periplasmic space in gram-negative bacteria. Using cryo-electron microscopy it has been found that a much smaller periplasmic space is also present in gram-positive bacteria, between cell wall and the plasma membrane. The periplasm may constitute up to 40% of the total cell volume of gram-negative bacteria, but is a much smaller percentage in gram-positive bacteria.
The ATP-binding cassette transporters are a transport system superfamily that is one of the largest and possibly one of the oldest gene families. It is represented in all extant phyla, from prokaryotes to humans. ABC transporters belong to translocases.

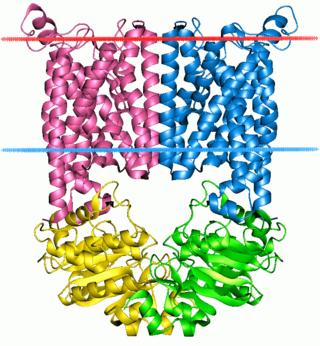
P-glycoprotein 1 also known as multidrug resistance protein 1 (MDR1) or ATP-binding cassette sub-family B member 1 (ABCB1) or cluster of differentiation 243 (CD243) is an important protein of the cell membrane that pumps many foreign substances out of cells. More formally, it is an ATP-dependent efflux pump with broad substrate specificity. It exists in animals, fungi, and bacteria, and it likely evolved as a defense mechanism against harmful substances.
In microbiology, efflux is the moving of a variety of different compounds out of cells, such as antibiotics, heavy metals, organic pollutants, plant-produced compounds, quorum sensing signals, bacterial metabolites and neurotransmitters. All microorganisms, with a few exceptions, have highly conserved DNA sequences in their genome that encode efflux pumps. Efflux pumps actively move substances out of a microorganism, in a process known as active efflux, which is a vital part of xenobiotic metabolism. This active efflux mechanism is responsible for various types of resistance to bacterial pathogens within bacterial species - the most concerning being antibiotic resistance because microorganisms can have adapted efflux pumps to divert toxins out of the cytoplasm and into extracellular media.

Cross-resistance is when something develops resistance to several substances that have a similar mechanism of action. For example, if a certain type of bacteria develops resistance to one antibiotic, that bacteria will also have resistance to several other antibiotics that target the same protein or use the same route to get into the bacterium. A real example of cross-resistance occurred for nalidixic acid and ciprofloxacin, which are both quinolone antibiotics. When bacteria developed resistance to ciprofloxacin, they also developed resistance to nalidixic acid because both drugs inhibit topoisomerase, a key enzyme in DNA replication. Due to cross-resistance, antimicrobial treatments like phage therapy can quickly lose their efficacy against bacteria. This makes cross-resistance an important consideration in designing evolutionary therapies.
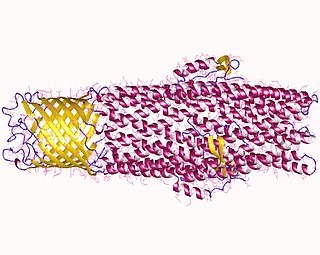
Proteins in the outer membrane efflux protein family form trimeric (three-piece) channels that allow export of a variety of substrates in gram-negative bacteria. Each member of this family is composed of two repeats. The trimeric channel is composed of a 12-stranded beta-barrel that spans the outer membrane, and a long all helical barrel that spans the periplasm.

Multidrug and toxin extrusion protein 1 (MATE1), also known as solute carrier family 47 member 1, is a protein that in humans is encoded by the SLC47A1 gene. SLC47A1 belongs to the MATE family of transporters that are found in bacteria, archaea and eukaryotes.

Multidrug and toxin extrusion protein 2 is a protein which in humans is encoded by the SLC47A2 gene.
Multi-antimicrobial extrusion protein (MATE) also known as multidrug and toxin extrusion or multidrug and toxic compound extrusion is a family of proteins which function as drug/sodium or proton antiporters.
SmeT is a transcriptional repressor protein of 24.6 kDa, found in the pathogenic bacteria Stenotrophomonas maltophilia. SmeT is responsible for the regulation of the Multidrug Resistance (MDR) efflux pump, SmeDEF, that gives the bacteria resistance to several antibiotics including macrolides, TMP/SMX, tetracycline, chloramphenicol, quinolones and erythromycin. SmeT is encoded 223 bp upstream of SmeDEF, with just 56 base pairs between their transcription start sites and an overlapping region between the promoters. The production of the SmeT protein downregulates its own transcription, along with that of the efflux pump by sterically hindering the binding of RNA Polymerase to the DNA. SmeDEF was the first MDR pump discovered in the S. maltophilia species. The pump is named by its different parts: SmeE, the transporter itself that spans the plasma membrane, SmeF, the protein on the outer portion of the membrane, and SmeD, a membrane fusion protein. On general purpose media and no selectors, the genes for MDR pumps are typically not expressed, and the repressor is found bound to the DNA. In fact, mutations in SmeT that lead to overexpression of SmeDEF can pose fitness challenges to the bacteria. However, this overexpression has been identified in the bacterium and may pose a threat to our health.
Plasmid-mediated resistance is the transfer of antibiotic resistance genes which are carried on plasmids. Plasmids possess mechanisms that ensure their independent replication as well as those that regulate their replication number and guarantee stable inheritance during cell division. By the conjugation process, they can stimulate lateral transfer between bacteria from various genera and kingdoms. Numerous plasmids contain addiction-inducing systems that are typically based on toxin-antitoxin factors and capable of killing daughter cells that don't inherit the plasmid during cell division. Plasmids often carry multiple antibiotic resistance genes, contributing to the spread of multidrug-resistance (MDR). Antibiotic resistance mediated by MDR plasmids severely limits the treatment options for the infections caused by Gram-negative bacteria, especially family Enterobacteriaceae. The global spread of MDR plasmids has been enhanced by selective pressure from antimicrobial medications used in medical facilities and when raising animals for food.
Cation diffusion facilitators (CDFs) are transmembrane proteins that provide tolerance of cells to divalent metal ions, such as cadmium, zinc, and cobalt. These proteins are considered to be efflux pumps that remove these divalent metal ions from cells. However, some members of the CDF superfamily are implicated in ion uptake. All members of the CDF family possess six putative transmembrane spanners with strongest conservation in the four N-terminal spanners. The Cation Diffusion Facilitator (CDF) Superfamily includes the following families:

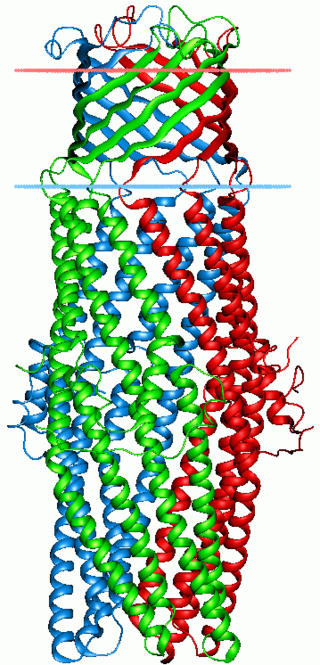
The Escherichia coliAcriflavine resistance encode a multi-drug efflux system that is believed to protect the bacterium against hydrophobic inhibitors. The E. coli AcrB protein is a transporter that is energized by proton-motive force and that shows the widest substrate specificity among all known multidrug pumps, ranging from most of the currently used antibiotics, disinfectants, dyes, and detergents to simple solvents.
The multidrug/oligosaccharidyl-lipid/polysaccharide (MOP) flippase superfamily is a group of integral membrane protein families. The MOP flippase superfamily includes twelve distantly related families, six for which functional data are available:
- One ubiquitous family (MATE) specific for drugs - (TC# 2.A.66.1) The Multi Antimicrobial Extrusion (MATE) Family
- One (PST) specific for polysaccharides and/or their lipid-linked precursors in prokaryotes - (TC# 2.A.66.2) The Polysaccharide Transport (PST) Family
- One (OLF) specific for lipid-linked oligosaccharide precursors of glycoproteins in eukaryotes - (TC# 2.A.66.3) The Oligosaccharidyl-lipid Flippase (OLF) Family
- One (MVF) lipid-peptidoglycan precursor flippase involved in cell wall biosynthesis - (TC# 2.A.66.4) The Mouse Virulence Factor (MVF) Family
- One (AgnG) which includes a single functionally characterized member that extrudes the antibiotic, Agrocin 84 - (TC# 2.A.66.5) The Agrocin 84 Antibiotic Exporter (AgnG) Family
- And finally, one (Ank) that shuttles inorganic pyrophosphate (PPi) - (TC# 2.A.66.9) The Progressive Ankylosis (Ank) Family
The Disulfide bond oxidoreductase D (DsbD) family is a member of the Lysine Exporter (LysE) Superfamily. A representative list of proteins belonging to the DsbD family can be found in the Transporter Classification Base.
Arsenite resistance (Ars) efflux pumps of bacteria may consist of two proteins, ArsB and ArsA, or of one protein. ArsA proteins have two ATP binding domains and probably arose by a tandem gene duplication event. ArsB proteins all possess twelve transmembrane spanners and may also have arisen by a tandem gene duplication event. Structurally, the Ars pumps resemble ABC-type efflux pumps, but there is no significant sequence similarity between the Ars and ABC pumps. When only ArsB is present, the system operates by a pmf-dependent mechanism, and consequently belongs in TC subclass 2.A. When ArsA is also present, ATP hydrolysis drives efflux, and consequently the system belongs in TC subclass 3.A. ArsB therefore appears twice in the TC system but ArsA appears only once. These pumps actively expel both arsenite and antimonite.
The 6TMS Neutral Amino Acid Transporter (NAAT) Family is a family of transporters belonging to the Lysine Exporter (LysE) Superfamily. Homologues are found in numerous Gram-negative and Gram-positive bacteria including many human pathogens. Several archaea also encode MarC homologues. Some of these organisms have 2 or more paralogues. Most of these proteins are of about the same size although a few are larger. They exhibit 6 putative TMSs. A representative list of members belonging to the NAAT family can be found in the Transporter Classification Database.
Multidrug resistance pumps also known Multidrug efflux pumps are a type of efflux pump and P-glycoprotein. MDR pumps in the cell membrane extrudes many foreign substances out of the cells and some pumps can have a broad specificity. MDR pumps exist in animals, fungi, and bacteria and likely evolved as a defense mechanism against harmful substances. There are seven families of MDRs and are grouped by homology, energy source, and overall structure.

Colin Hughes PhD ScD FLSW is a British microbiologist who has worked in the areas of bacterial virulence, motility and antibiotic resistance. He is Emeritus Professor of Microbiology at the University of Cambridge, Fellow of Trinity College Cambridge, and Fellow of the Learned Society of Wales.

Multidrug-resistant bacteria are bacteria that are resistant to three or more classes of antimicrobial drugs. MDR bacteria have seen an increase in prevalence in recent years and pose serious risks to public health. MDR bacteria can be broken into 3 main categories: Gram-positive, Gram-negative, and other (acid-stain). These bacteria employ various adaptations to avoid or mitigate the damage done by antimicrobials. With increased access to modern medicine there has been a sharp increase in the amount of antibiotics consumed. Given the abundant use of antibiotics there has been a considerable increase in the evolution of antimicrobial resistance factors, now outpacing the development of new antibiotics.












