
In molecular biology and genetics, transformation is the genetic alteration of a cell resulting from the direct uptake and incorporation of exogenous genetic material from its surroundings through the cell membrane(s). For transformation to take place, the recipient bacterium must be in a state of competence, which might occur in nature as a time-limited response to environmental conditions such as starvation and cell density, and may also be induced in a laboratory.

Agrobacterium tumefaciens is the causal agent of crown gall disease in over 140 species of eudicots. It is a rod-shaped, Gram-negative soil bacterium. Symptoms are caused by the insertion of a small segment of DNA, from a plasmid into the plant cell, which is incorporated at a semi-random location into the plant genome. Plant genomes can be engineered by use of Agrobacterium for the delivery of sequences hosted in T-DNA binary vectors.

Pseudomonas putida is a Gram-negative, rod-shaped, saprophytic soil bacterium. It has a versatile metabolism and is amenable to genetic manipulation, making it a common organism used in research, bioremediation, and synthesis of chemicals and other compounds.
Thermus thermophilus is a Gram-negative bacterium used in a range of biotechnological applications, including as a model organism for genetic manipulation, structural genomics, and systems biology. The bacterium is extremely thermophilic, with an optimal growth temperature of about 65 °C (149 °F). Thermus thermophilus was originally isolated from a thermal vent within a hot spring in Izu, Japan by Tairo Oshima and Kazutomo Imahori. The organism has also been found to be important in the degradation of organic materials in the thermogenic phase of composting. T. thermophilus is classified into several strains, of which HB8 and HB27 are the most commonly used in laboratory environments. Genome analyses of these strains were independently completed in 2004. Thermus also displays the highest frequencies of natural transformation known to date.
Paenarthrobacter ureafaciens KI72, popularly known as nylon-eating bacteria, is a strain of Paenarthrobacter ureafaciens that can digest certain by-products of nylon 6 manufacture. It uses a set of enzymes to digest nylon, popularly known as nylonase.

Rhodococcus is a genus of aerobic, nonsporulating, nonmotile Gram-positive bacteria closely related to Mycobacterium and Corynebacterium. While a few species are pathogenic, most are benign, and have been found to thrive in a broad range of environments, including soil, water, and eukaryotic cells. Some species have large genomes, including the 9.7 megabasepair genome of Rhodococcus sp. RHA1.
Pseudomonas citronellolis is a Gram-negative, bacillus bacterium that is used to study the mechanisms of pyruvate carboxylase. It was first isolated from forest soil, under pine trees, in northern Virginia, United States.
Pseudomonas veronii is a Gram-negative, rod-shaped, fluorescent, motile bacterium isolated from natural springs in France. It may be used for bioremediation of contaminated soils, as it has been shown to degrade a variety of simple aromatic organic compounds. Based on 16S rRNA analysis, P. veronii has been placed in the P. fluorescens group.
Pseudomonas stutzeri is a Gram-negative soil bacterium that is motile, has a single polar flagellum, and is classified as bacillus, or rod-shaped. While this bacterium was first isolated from human spinal fluid, it has since been found in many different environments due to its various characteristics and metabolic capabilities. P. stutzeri is an opportunistic pathogen in clinical settings, although infections are rare. Based on 16S rRNA analysis, this bacterium has been placed in the P. stutzeri group, to which it lends its name.
Microbial biodegradation is the use of bioremediation and biotransformation methods to harness the naturally occurring ability of microbial xenobiotic metabolism to degrade, transform or accumulate environmental pollutants, including hydrocarbons, polychlorinated biphenyls (PCBs), polyaromatic hydrocarbons (PAHs), heterocyclic compounds, pharmaceutical substances, radionuclides and metals.

Rhodococcus equi is a Gram-positive coccobacillus bacterium. The organism is commonly found in dry and dusty soil and can be important for diseases of domesticated animals. The frequency of infection can reach near 60%. R. equi is an important pathogen causing pneumonia in foals. Since 2008, R. equi has been known to infect wild boar and domestic pigs. R. equi can infect immunocompromised people, such as HIV-AIDS patients or transplant recipients. Rhodococcus infection in these patients resemble clinical and pathological signs of pulmonary tuberculosis. It is a facultative intracellular mycobacterial pathogen.
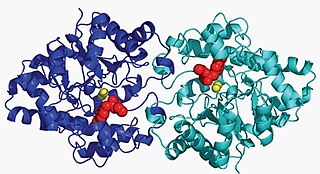
Aryldialkylphosphatase is a metalloenzyme that hydrolyzes the triester linkage found in organophosphate insecticides:

Bacillus anthracis is a gram-positive and rod-shaped bacterium that causes anthrax, a deadly disease to livestock and, occasionally, to humans. It is the only permanent (obligate) pathogen within the genus Bacillus. Its infection is a type of zoonosis, as it is transmitted from animals to humans. It was discovered by a German physician Robert Koch in 1876, and became the first bacterium to be experimentally shown as a pathogen. The discovery was also the first scientific evidence for the germ theory of diseases.
Cupriavidus metallidurans is a non-spore-forming, Gram-negative bacterium which is adapted to survive several forms of heavy metal stress.
Rhodopseudomonas palustris is a rod-shaped, Gram-negative purple nonsulfur bacterium, notable for its ability to switch between four different modes of metabolism.
Sphingomonas yanoikuyae is a short rod-shaped, strictly aerobic, Gram-negative, non-motile, non-spore-forming, chemoheterotrophic species of bacteria that is yellow or off-white in color. Its type strain is JCM 7371. It is notable for degrading a variety of aromatic compounds including biphenyl, naphthalene, phenanthrene, toluene, m-, and p-xylene. S. yanoikuyae was discovered by Brian Goodman on the southern coast of Papua New Guinea. However, Sphingomonas have a wide distribution across freshwater, seawater, and terrestrial habitats. This is due to the bacteria's ability to grow and survive under low-nutrient conditions as it can utilize a broad range of organic compounds.
Desulfitobacterium hafniense is a species of gram positive bacteria, its type strain is DCB-2T..
Rhodococcus opacus is a bacterium species in the genus Rhodococcus. It is moderately chemolithotrophic. Its genome has been sequenced. R. opacus possesses extensive catabolic pathways for both sugars and aromatics and can tolerate inhibitory compounds found in depolymerized biomass

Corynebacterium xerosis is a Gram-positive, rod-shaped bacterium in the genus Corynebacterium. Although it is frequently a harmless commensal organism living on the skin and mucus membranes, C. xerosis is also a clinically-relevant opportunistic pathogen that has been attributed to a number of different infections in animals and humans. However, its actual prominence in human medicine is up for debate due to early difficulties distinguishing it from other Corynebacterium species in clinical isolates.

The species Rhizorhabdus wittichii, formerly Sphingomonas wittichii, is a Gram-negative, rod-shaped motile bacterium, with an optimum growth temperature at 30 °C. It forms a greyish white colony. It has been found to have a 67 mol% of DNA G+C content.









